Example of practical application of copulas
In a previous article, I described the theoretical rationale for copulas. Since I myself was a student, I know that the best explanation of the theoretical apparatus can be an example of its practical application. Therefore, in this article I will try to show how copulas are used to model the interdependencies of several random variables.

As already mentioned, many copulas can reflect the dependence of not only two, but several values at once. This, of course, is good, but standard copulas imply some symmetry, that is, all pairwise dependencies will have the same pattern. Agree, this is not always true. To overcome this problem, an interesting and quite obvious (how much torment is hidden behind this word in mathematical analysis) method was proposed - we divide the joint copula into a set of two-dimensional copulas. We know that for n variables we will have pairwise relationships. That is, we will have to determine just so many pairwise copulas. We show that the joint distribution density of 4 quantities can be expressed as follows:
pairwise relationships. That is, we will have to determine just so many pairwise copulas. We show that the joint distribution density of 4 quantities can be expressed as follows:
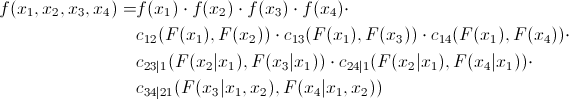
There are several ways to represent the partition, and the above is called the canonical vine (I sincerely apologize, but I did not find the established translation of “canonical vine” in the mathematical context, so I will use the direct translation). Graphically, the last equation can be represented as follows:

Three trees describe a partition of a 4-dimensional copula. In the leftmost tree, circles are marginal distributions, arrows are two-dimensional copulas. Our task, as the person responsible for modeling, is to set all the “arrows”, that is, to determine pairwise copulas and their parameters.
Now let's look at the formula of the D-shaped vine:
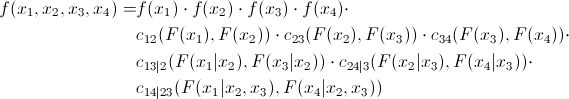
and its graphical representation
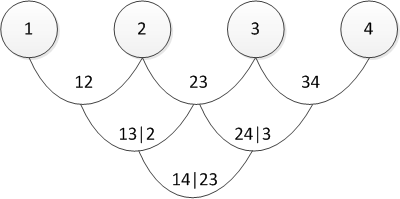
In fact, the canonical and D-vines differ only in the order in which pairwise dependencies are determined. One may ask, “how will I choose the order?” Or the equivalent “what vine should I choose?” I have an answer to this: in practice, they start with pairs that show the greatest relationship. The latter is determined by Kendall's tau . Do not worry - everything will become clearer later, when I will show a direct practical example.
After you have decided on the order of definition of the copula, you need to understand which copula to use to determine the connections (Gauss, student, Gumbel or what else). In principle, there is no exact scientific method. Someone looks at paired graphs of the distribution of “archived” quantities and determine the copula by eye. Someone is using Chi charts (it says how to use them ). A matter of mastery and a vast field for research.
After we decide which copulas to use in pair relationships, we need to determine their parameters. This is done by the numerical maximum likelihood method (numerical MLE) using the following algorithms:
These are algorithms for calculating the logarithmic likelihood function, which, as you know, needs to be maximized. This is a task for various optimization packages. Here, a little explanation is required on the variables used in the "code":
x is a set of "archived" data of size T by n, where x is bounded on the interval (0,1).
v is a matrix of size T by n-1 by n-1, where intermediate data will be loaded.
param - parameters of paired copulas, which we are looking for, maximizing Log-likelihood.
L (x, v, param) =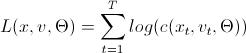
h (x, v, param) =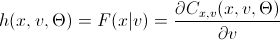
Lovers of spoiling the paper with their calculations may not open the next spoiler, where I introduced the h-functions of the main copulas.
After we find the parameters of our copulas that maximize the logarithmic likelihood function, we can proceed to generate random variables that have the same relationship structure as our initial data. For this, there are also algorithms:
You use the parameters that were obtained using the model estimation algorithms and generate n variables. Run this algorithm T times and get the matrix T by n, the data in which have the same relationship pattern as the original data. The only innovation compared to the previous algorithms is the inverse h-function (h_inv), which is the inverse function of the conditional distribution over the first variable.
Again, let me save you from lengthy calculations:
That's basically it. And now the promised ...
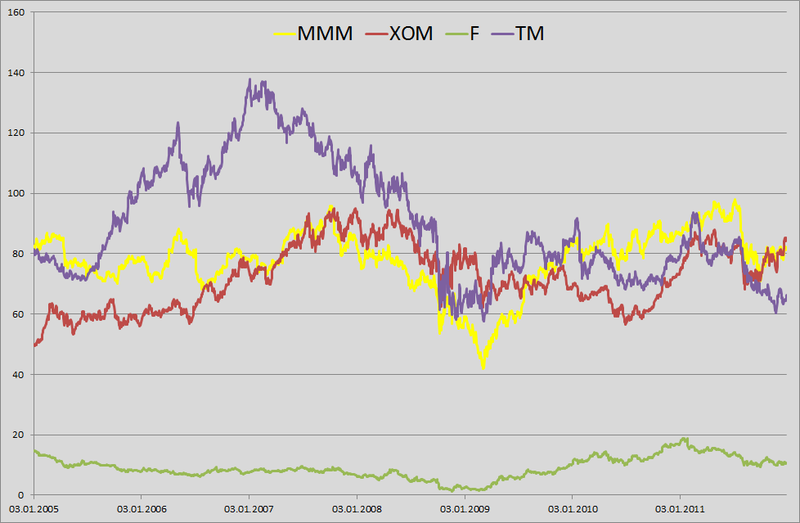
1. For example, I decided to play a small bank clerk who was told to calculate the daily VaR for his portfolio. In the yard on January 1, 2012 and in my portfolio in equal shares are the shares of 4 companies: 3M (MMM), Exxon Mobil (XOM), Ford Motors (F) and Toyota Motors (TM). Having pestered with good ah-tishniks, getting them to unload price history from our server for the last 6 years, I realized that they had no time (January 1 in the yard, who works at all ???) and unloaded this information from Yahu's server. I took a daily price history from January 3, 2005 to December 31, 2011. The matrix was 1763 by 4.
2. I found the log-retar: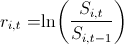 and built their pairwise distribution.
and built their pairwise distribution.
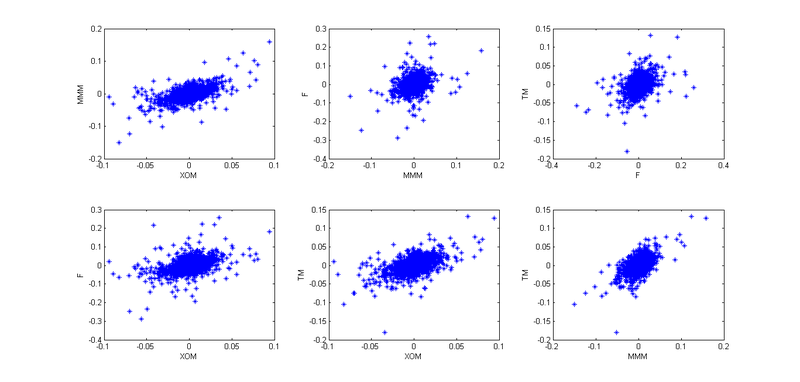
Well, to present the big picture.
3. Then I remembered the lectures on finance and how we were hammered into us - “guys, the fact that the retarns are distributed normally is rubbish; in real life, this is not so. ” I thought if I should use the assumption that the retarns are distributed according to NIG (Normal Inverse Gaussian). Studies show that this distribution is in good agreement with empirical data. But I need to come up with a VaR calculation model, and then test it on historical data. Any model that does not take into account the clustering of the second moment will show bad results. Then the idea came - I will use GARCH (1,1). The model grasps 3 and 4 moments of distribution quite well, plus takes into account the volatility of the second moment. So I decided that my reters follow the following law . Here with - is the historical average, the z - a standard normally distributed random variable, and the sigma should have their law:
. Here with - is the historical average, the z - a standard normally distributed random variable, and the sigma should have their law: 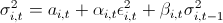 .
.
4. So, now we need to take into account the interdependence of these four stocks. To begin with, I used empirical copulas and plotted the pair dependencies of the “archived” observations:
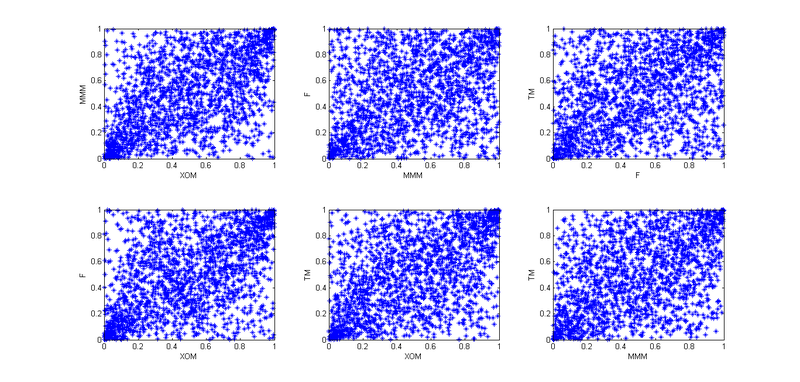
Given that today is January 1, and I am a lazy cattle, I decided that all the patterns here are similar to Student’s copula, but in order to better convey the interdependencies, I decided not to use 4- x dimensional copula, and break it all into six 2-dimensional copulas.
5. In order to split into paired copulas, you need to choose the presentation order - canonical or D-vine. For this, I counted Kendall's tau to determine the level of interdependence:
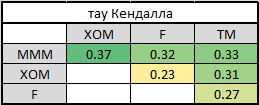
It is easy to see that MMM has the strongest ties with all other companies. This determines our choice of vines - canonical. Why? Press PageUp a couple of times and look at its graphical display. In the first tree, all arrows come from one node - this is exactly our situation when one company has the strongest connections. If Kendall’s high tau in the last table were on different lines and different columns, we would choose a D-vine.
6. So we have determined the order of presentation and all the necessary pairwise copulas - we can begin to evaluate the parameters. Applying the above algorithm for evaluating the parameters of the canonical vine, I found all the necessary parameters.
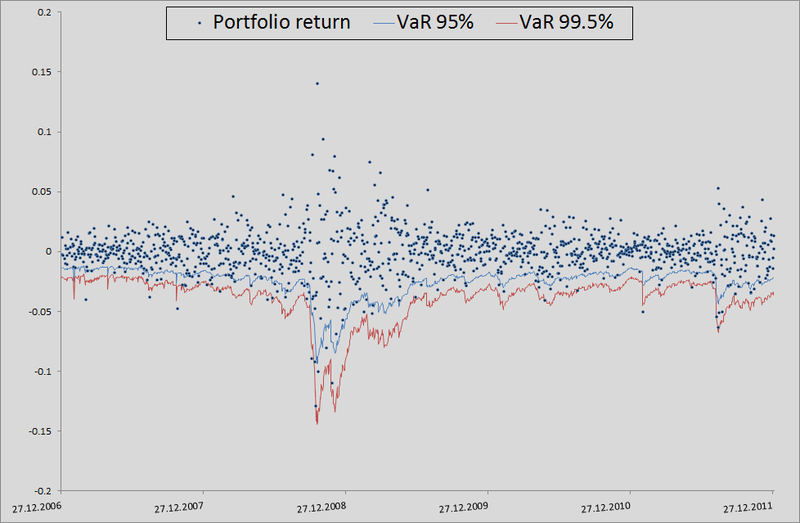
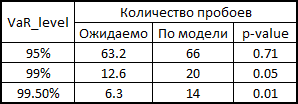
7. Here I will explain a little how this all happened in the context of backtesting. In order to check how plausibly my model considers VaR, I wanted to see how it handled in the past. To do this, I gave her a small head start - 500 days and, starting from 501 days, I considered my risk metrics for the next day. Having the parameters of the copula on hand, I generated 10,000 fours of evenly distributed values, obtained from them fours of normally distributed values (noise) and substituted into the formula for calculating the retrograde from 3 points of this example. There I inserted a sigma forecast by the GARCH (1,1) model using data from previous days (this was done by a combination of the Matlab functions ugarch and ugarchpred) and historical averages. Having thus received 10,000 fours of the stocks of stocks, I received 10,000 of the stocks of my portfolio,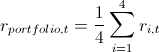 (since we have equal shares and log-reters, we have the right to do so). Then sorting and highlighting of the 50th, 100th and 500th values for 99.5%, 99% and 95% of the threshold level, respectively. Having thus calculated the risk of the metric for the remaining 1263 days, I got a beautiful graph and statistics of breakdowns of levels.
(since we have equal shares and log-reters, we have the right to do so). Then sorting and highlighting of the 50th, 100th and 500th values for 99.5%, 99% and 95% of the threshold level, respectively. Having thus calculated the risk of the metric for the remaining 1263 days, I got a beautiful graph and statistics of breakdowns of levels.
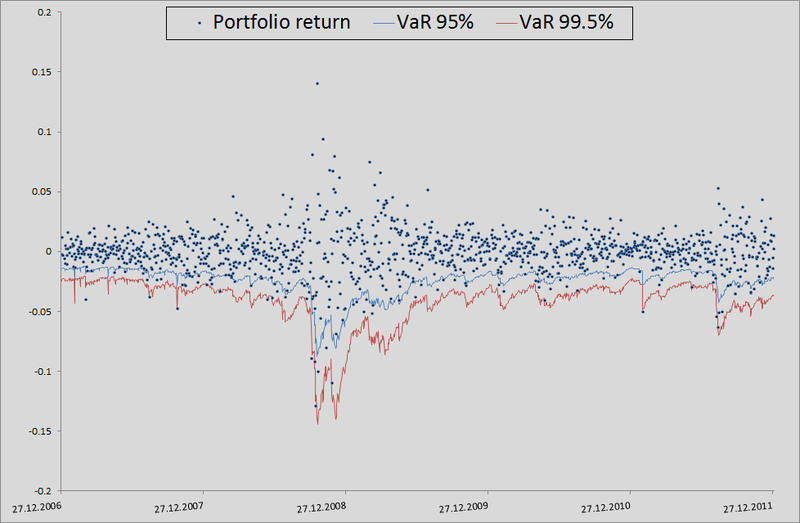
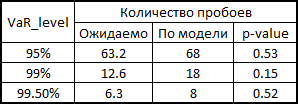
In principle, there is still much to grow in accuracy. But if we take the model as a benchmark without taking into account interdependencies in general, then there is a seam in general:
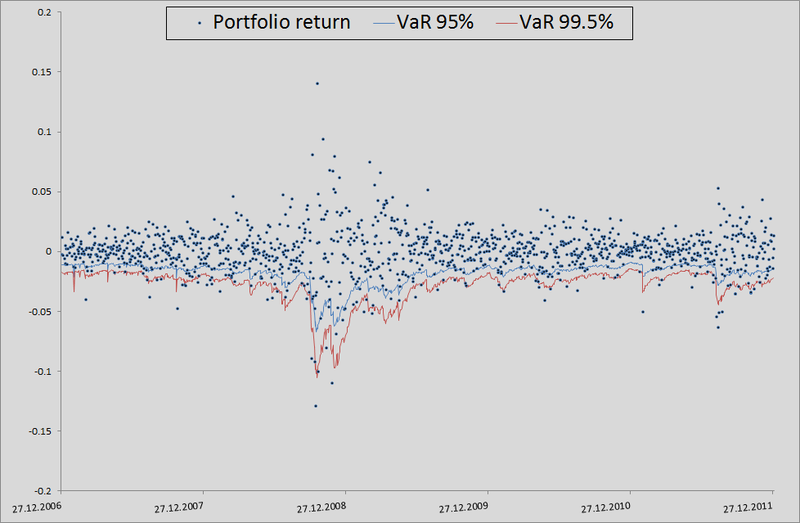
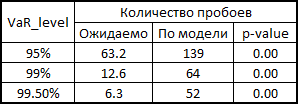
To be honest, we need to show how the model would behave if we used correlations and generated noise to calculate the retrens from the multidimensional normal distribution:
As you can see, for VaR 95 % this model is even better than ours. But the farther, the worse - as already mentioned, the normal distribution does not catch the tails.
In general, copulas can be used, but this must be done wisely. They give wider possibilities for taking into account interdependencies, but, as with any exact model, one must be careful here - it is possible to miss the assumptions and get results worse than from the standard model.
PS I have often noticed tongue-tied behind me, so I will be glad if you point me to such moments in the text. I tried to express my thoughts as clearly as possible, but not always I succeed.

Again a little theory
As already mentioned, many copulas can reflect the dependence of not only two, but several values at once. This, of course, is good, but standard copulas imply some symmetry, that is, all pairwise dependencies will have the same pattern. Agree, this is not always true. To overcome this problem, an interesting and quite obvious (how much torment is hidden behind this word in mathematical analysis) method was proposed - we divide the joint copula into a set of two-dimensional copulas. We know that for n variables we will have
 pairwise relationships. That is, we will have to determine just so many pairwise copulas. We show that the joint distribution density of 4 quantities can be expressed as follows:
pairwise relationships. That is, we will have to determine just so many pairwise copulas. We show that the joint distribution density of 4 quantities can be expressed as follows: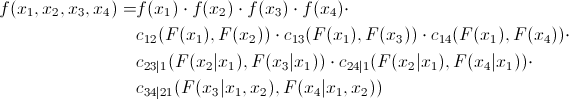
little evidence
To begin with, we recall that the distribution density of several quantities can be expressed as follows:

Also from the definition of a copula, we know that:

which in turn can be expressed as follows:

For a two-dimensional case, it will look like this:

And combining this with the very first formula , we get:

You can expand this example to the three-dimensional case:

and combining with the previous formula, we get:

Thus, we see that all elements from the first equation can be represented as a product of paired copulas and marginal oh density.

Also from the definition of a copula, we know that:

which in turn can be expressed as follows:

For a two-dimensional case, it will look like this:

And combining this with the very first formula , we get:

You can expand this example to the three-dimensional case:

and combining with the previous formula, we get:

Thus, we see that all elements from the first equation can be represented as a product of paired copulas and marginal oh density.
There are several ways to represent the partition, and the above is called the canonical vine (I sincerely apologize, but I did not find the established translation of “canonical vine” in the mathematical context, so I will use the direct translation). Graphically, the last equation can be represented as follows:

Three trees describe a partition of a 4-dimensional copula. In the leftmost tree, circles are marginal distributions, arrows are two-dimensional copulas. Our task, as the person responsible for modeling, is to set all the “arrows”, that is, to determine pairwise copulas and their parameters.
Now let's look at the formula of the D-shaped vine:
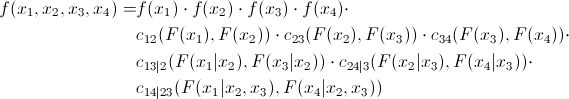
and its graphical representation

In fact, the canonical and D-vines differ only in the order in which pairwise dependencies are determined. One may ask, “how will I choose the order?” Or the equivalent “what vine should I choose?” I have an answer to this: in practice, they start with pairs that show the greatest relationship. The latter is determined by Kendall's tau . Do not worry - everything will become clearer later, when I will show a direct practical example.
After you have decided on the order of definition of the copula, you need to understand which copula to use to determine the connections (Gauss, student, Gumbel or what else). In principle, there is no exact scientific method. Someone looks at paired graphs of the distribution of “archived” quantities and determine the copula by eye. Someone is using Chi charts (it says how to use them ). A matter of mastery and a vast field for research.
After we decide which copulas to use in pair relationships, we need to determine their parameters. This is done by the numerical maximum likelihood method (numerical MLE) using the following algorithms:
Pseudocode for evaluating canonical vine parameters
Log-likelihood=0;
for i = 1:n
v(0,i) = x(i)
end for
for j = 1:n-1
for i = 1:n-j
Log-likelihood = Log-likelihood + L(v(j-1,1), v(j-1,i+1), param(j,i));
end for
if j == n then
break
end if
for i = 1:n-j
v(j,i) = h(v(j-1,i+1), v(j-1,1), param(j,i));
end for
end for
Pseudocode for evaluating D-vine parameters
Log-likelihood=0;
for i = 1:n
v(0,i) = x(i);
end for
for j = 1:n-1
Log-likelihood = Log-likelihood + L(v(0,i),v(0,i+1), param(1,i));
end for
v(1,1) = h(v(0,1),v(0,2), param(1,1))
for k = 1:n-3
v(1,2k) = h(v(0,k+2), v(0,k+1), param(1,k+1));
v(1,2k+1) = h(v(0,k+1), v(0,k+2), param(1,k+1));
end for
v(1,2n-4) = h(v(0,n), v(0,n-1), param(1,n-1));
for j = 2:n-1
for i = 1:n-j
Log-likelihood = Log-likelihood + L(v(j-1,2i-1),v(j-1,2i), param(j,i));
end for
if j == n-1 then
break
end if
v(j,i) = h(v(j-1,1), v(j-1,2), param(j,1));
if n > 4 then
for i = 1:n-j-2
v(j,2i) = h(v(j-1,2i+2),v(j-1,2i+1),param(j,i+1));
v(j,2i+1) = h(v(j-1,2i+1),v(j-1,2i+2),param(j,i+1));
end for
end if
v(j,2n-2j-2) = h(v(j-1,2n-2j),v(j-1,2n-2j-1),param(j,n-j))
end for
These are algorithms for calculating the logarithmic likelihood function, which, as you know, needs to be maximized. This is a task for various optimization packages. Here, a little explanation is required on the variables used in the "code":
x is a set of "archived" data of size T by n, where x is bounded on the interval (0,1).
v is a matrix of size T by n-1 by n-1, where intermediate data will be loaded.
param - parameters of paired copulas, which we are looking for, maximizing Log-likelihood.
L (x, v, param) =
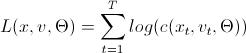
h (x, v, param) =
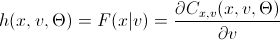
Lovers of spoiling the paper with their calculations may not open the next spoiler, where I introduced the h-functions of the main copulas.
h functions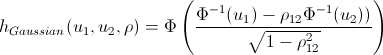
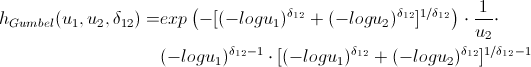
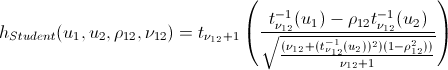
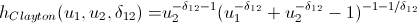
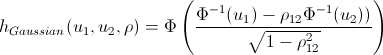
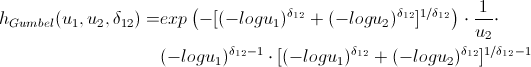
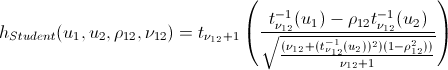
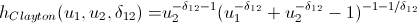
After we find the parameters of our copulas that maximize the logarithmic likelihood function, we can proceed to generate random variables that have the same relationship structure as our initial data. For this, there are also algorithms:
Pseudocode for generating canonical vine data
w = rand(n,1); % Генерируем n равномерно распределенных величин
x(1) = v(1,1) = w(1);
for i = 2:n
v(i,1) = w(i);
for k = i-1:1
v(i,1) = h_inv(v(i,1), v(k,k), param(k,i-k)); % стоит отметить, что здесь идет обратный цикл.
end for
x(i) = v(i,1);
if i = n then
break
end if
for j = 1:i-1
v(i,j+1) = h(v(i,j), v(j,j), param(j,i-j));
end for
end for
Pseudo-code for generating D-vine data
w = rand(n,1); % Генерируем n равномерно распределенных величин
x(1) = v(1,1) = w(1);
x(2) = v(2,1) = h_inv(w(2), v(1,1), param(1,1));
v(2,2) = h(v(1,1), v(2,1), param(1,1));
for i = 3:n
v(i,1) = w(i);
for k = i-1:2
v(i,1) = h_inv(v(i,1), v(i-1,2k-2), param(k,i-k));
end for
v(i,1) = h_inv(v(i,1), v(i-1,1), param(k,i-1));
x(i) = v(i,1);
if i == n
break
end if
v(i,2) = h(v(i-1,1), v(i,1), param(1,i-1));
v(i,3) = h(v(i,1), v(i-1,1), param(1,i-1));
if i>3
for j = 2:i-2
v(i,2j) = h(v(i-1,2j-2), v(i,2j-1), param(j,i-j));
v(i,2j+1) = h(v(i,2j-1), v(i-1,2j-2), param(j,i-j));
end for
end if
v(i,2i-2) = h(v(i-1,2i-4), v(i,2i-3), param(i-1,1));
end for
You use the parameters that were obtained using the model estimation algorithms and generate n variables. Run this algorithm T times and get the matrix T by n, the data in which have the same relationship pattern as the original data. The only innovation compared to the previous algorithms is the inverse h-function (h_inv), which is the inverse function of the conditional distribution over the first variable.
Again, let me save you from lengthy calculations:
inverse h-functions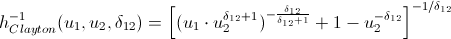
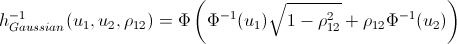
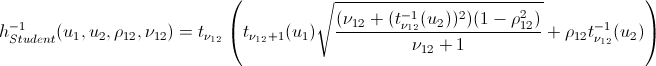
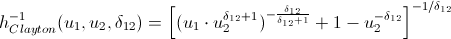
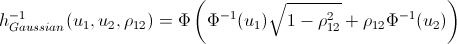
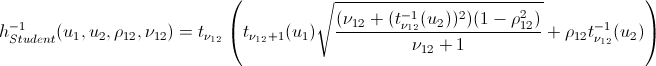
That's basically it. And now the promised ...
Practical example
1. For example, I decided to play a small bank clerk who was told to calculate the daily VaR for his portfolio. In the yard on January 1, 2012 and in my portfolio in equal shares are the shares of 4 companies: 3M (MMM), Exxon Mobil (XOM), Ford Motors (F) and Toyota Motors (TM). Having pestered with good ah-tishniks, getting them to unload price history from our server for the last 6 years, I realized that they had no time (January 1 in the yard, who works at all ???) and unloaded this information from Yahu's server. I took a daily price history from January 3, 2005 to December 31, 2011. The matrix was 1763 by 4.

2. I found the log-retar:
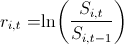 and built their pairwise distribution.
and built their pairwise distribution. 
Well, to present the big picture.
3. Then I remembered the lectures on finance and how we were hammered into us - “guys, the fact that the retarns are distributed normally is rubbish; in real life, this is not so. ” I thought if I should use the assumption that the retarns are distributed according to NIG (Normal Inverse Gaussian). Studies show that this distribution is in good agreement with empirical data. But I need to come up with a VaR calculation model, and then test it on historical data. Any model that does not take into account the clustering of the second moment will show bad results. Then the idea came - I will use GARCH (1,1). The model grasps 3 and 4 moments of distribution quite well, plus takes into account the volatility of the second moment. So I decided that my reters follow the following law
 . Here with - is the historical average, the z - a standard normally distributed random variable, and the sigma should have their law:
. Here with - is the historical average, the z - a standard normally distributed random variable, and the sigma should have their law: 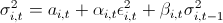 .
. 4. So, now we need to take into account the interdependence of these four stocks. To begin with, I used empirical copulas and plotted the pair dependencies of the “archived” observations:

Given that today is January 1, and I am a lazy cattle, I decided that all the patterns here are similar to Student’s copula, but in order to better convey the interdependencies, I decided not to use 4- x dimensional copula, and break it all into six 2-dimensional copulas.
5. In order to split into paired copulas, you need to choose the presentation order - canonical or D-vine. For this, I counted Kendall's tau to determine the level of interdependence:

It is easy to see that MMM has the strongest ties with all other companies. This determines our choice of vines - canonical. Why? Press PageUp a couple of times and look at its graphical display. In the first tree, all arrows come from one node - this is exactly our situation when one company has the strongest connections. If Kendall’s high tau in the last table were on different lines and different columns, we would choose a D-vine.
6. So we have determined the order of presentation and all the necessary pairwise copulas - we can begin to evaluate the parameters. Applying the above algorithm for evaluating the parameters of the canonical vine, I found all the necessary parameters.
7. Here I will explain a little how this all happened in the context of backtesting. In order to check how plausibly my model considers VaR, I wanted to see how it handled in the past. To do this, I gave her a small head start - 500 days and, starting from 501 days, I considered my risk metrics for the next day. Having the parameters of the copula on hand, I generated 10,000 fours of evenly distributed values, obtained from them fours of normally distributed values (noise) and substituted into the formula for calculating the retrograde from 3 points of this example. There I inserted a sigma forecast by the GARCH (1,1) model using data from previous days (this was done by a combination of the Matlab functions ugarch and ugarchpred) and historical averages. Having thus received 10,000 fours of the stocks of stocks, I received 10,000 of the stocks of my portfolio,
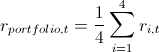 (since we have equal shares and log-reters, we have the right to do so). Then sorting and highlighting of the 50th, 100th and 500th values for 99.5%, 99% and 95% of the threshold level, respectively. Having thus calculated the risk of the metric for the remaining 1263 days, I got a beautiful graph and statistics of breakdowns of levels.
(since we have equal shares and log-reters, we have the right to do so). Then sorting and highlighting of the 50th, 100th and 500th values for 99.5%, 99% and 95% of the threshold level, respectively. Having thus calculated the risk of the metric for the remaining 1263 days, I got a beautiful graph and statistics of breakdowns of levels.

How to read p-value for VaR breakdown statistics
In 1995, Comrade Kupich proposed a formula for calculating the reliability of the VaR calculation model:

Here x is the number of breakdowns according to the model, T is the number of tests (we have = 1263), p is the expected share of breakdowns (in our case 0.5%, 1% and 5%).
According to the null hypothesis, these statistics have a chi-square distribution with one degree of freedom. Having found the statistics, it is easy to calculate its p-value.

Here x is the number of breakdowns according to the model, T is the number of tests (we have = 1263), p is the expected share of breakdowns (in our case 0.5%, 1% and 5%).
According to the null hypothesis, these statistics have a chi-square distribution with one degree of freedom. Having found the statistics, it is easy to calculate its p-value.
In principle, there is still much to grow in accuracy. But if we take the model as a benchmark without taking into account interdependencies in general, then there is a seam in general:


To be honest, we need to show how the model would behave if we used correlations and generated noise to calculate the retrens from the multidimensional normal distribution:


As you can see, for VaR 95 % this model is even better than ours. But the farther, the worse - as already mentioned, the normal distribution does not catch the tails.
In general, copulas can be used, but this must be done wisely. They give wider possibilities for taking into account interdependencies, but, as with any exact model, one must be careful here - it is possible to miss the assumptions and get results worse than from the standard model.
PS I have often noticed tongue-tied behind me, so I will be glad if you point me to such moments in the text. I tried to express my thoughts as clearly as possible, but not always I succeed.
