DNA Mechanisms for storing and processing information. Part I

A lot of people use the term DNA. But there are almost no articles describing how it works (not understandable to biologists). I have already described in general terms the structure of the cell and the very basics of its energy processes . We now turn to DNA.
DNA stores information. Everybody knows that. But how does she do it?
To begin with, where it is stored in a cage. Approximately 98% is stored in the kernel. The rest is in mitochondria and chloroplasts (photosynthesis takes place in these guys). DNA is a huge polymer composed of monomeric units. It looks like this.
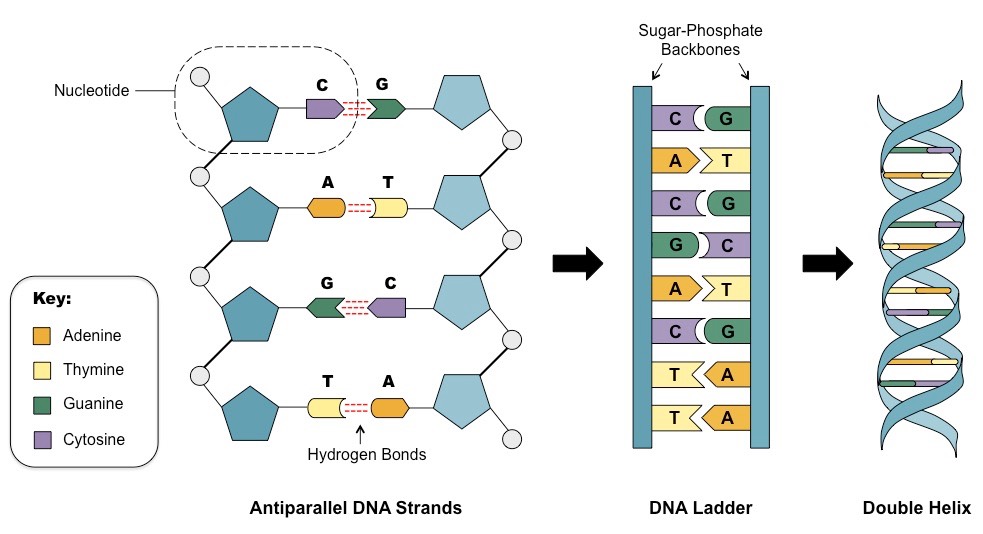
What do we see here? First, DNA is a double-stranded molecule. Why it is so important - a little later. Next we see the blue pentagons. These are deoxy molecules.ribose (such sugar, a little less glucose. It differs from ribose by the absence of one OH group, which gives stability to the DNA molecule, unlike RNA, which uses ribose. Next, for simplicity, I’ll omit the deoxy prefix and just say ribose, forgive us for scrupulous comrades). Small circles - residues of phosphoric acid. Well, actually there are nitrogen bases. There are 5 of them altogether, but in DNA there are mainly 4. These are Adenine, Guanin, Timine and Cytosine. That is, there is a ribose with which the nitrogenous base is associated. Together they form the so-called nucleosides, which bind to each other with the help of phosphoric acid residues. Thus we get a long chain consisting of monomers. Now look at the enlarged left chain. You see C and G are connected by three dashed lines, and T and A are two. What does it mean? Yes, DNA consists of two chains, but what keeps them together? There is such a thing as a hydrogen bond. It looks like this. A partial negative charge forms on the oxygen (O) and nitrogen (N) atoms, and a positive charge on the hydrogen (H). This leads to the formation of weak links.

The connections are really very weak. Their energy can be 200 times lower than the energy of covalent bonds (formed due to the overlapping of a pair of electron clouds, for example, a bond in a CO2 molecule). However, there are many such connections. In each of our cells, the DNA chains are connected by almost 16 billion weak links, not a few, do you agree?
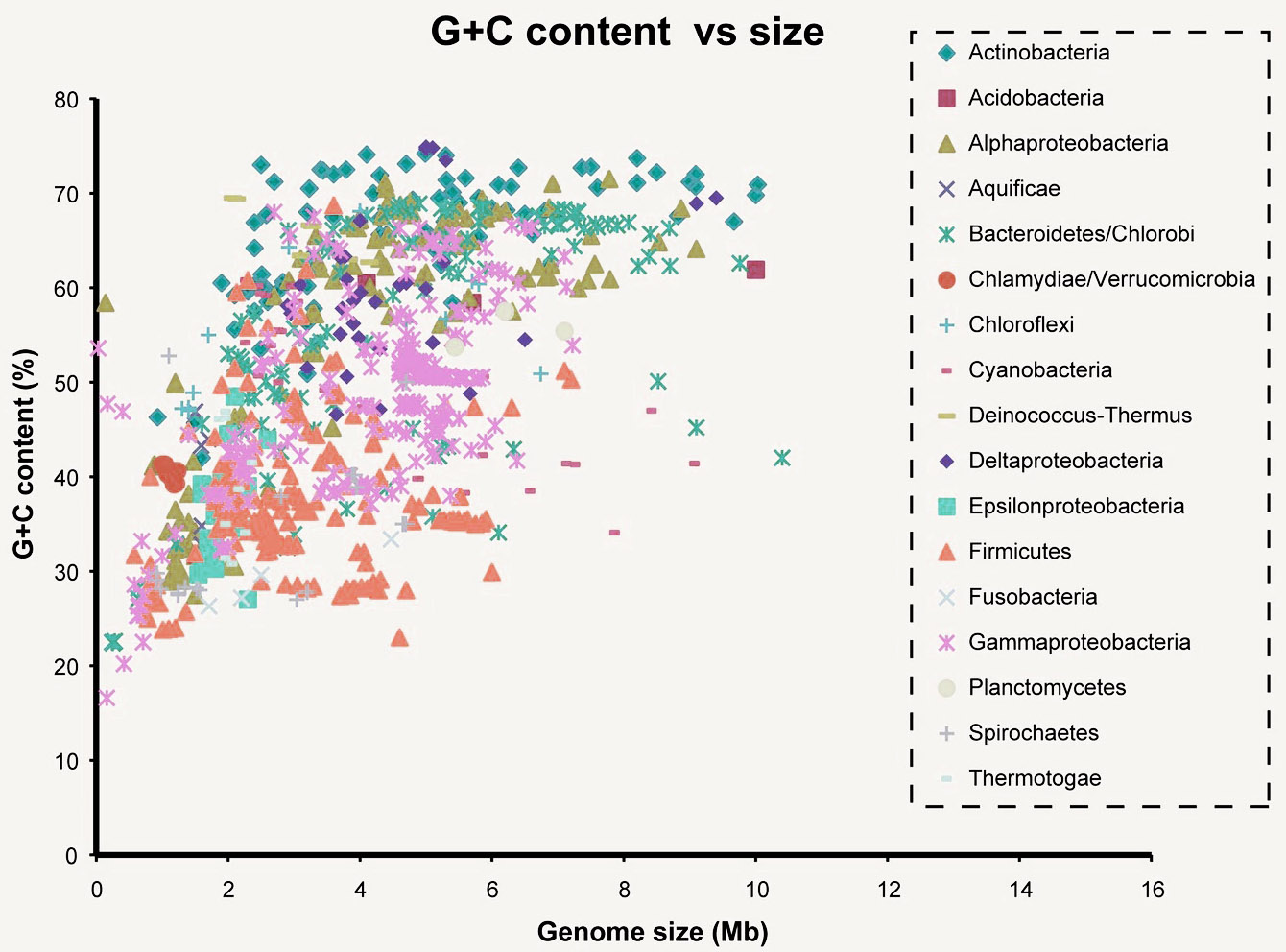
But back to the number of links between the bases. Cytosine and Guanine are connected by three connections, and Adenin and Tymin - by two. This leads to the fact that G and C are connected much stronger than A and T. Some organisms need special stability of DNA bonds, for example, living at high temperatures. When heated, DNA containing more HZ pairs is more stable. So you want to live in a geyser - have a lot of HZ pairs. Although recent studies say that there is no clear connection between the GC composition (% GC pairs from all pairs) and the temperature of the habitat. It should be said that it varies greatly. So, Candidatus Carsonella ruddii PV (intracellular endosymbiont) is about 16%, we have almost 41%, and Anaeromyxobacter K (the bacterium is quite average size) reaches 75%.
Here you can see the connection of the GC composition with the size of the bacterial genome. Mb is a million nucleotide pairs. The indicator is quite variable. By the way, it is often used as a feature when teaching various kinds of classifiers. He himself recently wrote a classifier for recognizing pathogens based on raw sequencing data and it turned out that the GC composition can be used even for one reader.
Until I forgot. Why is it important that DNA is double stranded? On the basis of one circuit, you can restore another. If in one chain there is a damaged piece opposite to the sequence Adenine-Adenine-Cytosine, then we know for sure that before the damage there was Timing-Timin-Guanin. Thus, the presence of the second circuit allows you to store information more reliably.
Cool! Now back to the DNA molecule itself. This is a chain of 4 types of links. But how long? Candidatus Carsonella ruddii PV, already mentioned above, has a total of 160,000 nucleotides. We have 3.2 billion with you (in a haploid cell, that is, with one set of chromosomes. Most of our cells have two). It seems a lot, right? Not really. In the unicellular amoeba (Amoeba dubia), it is approximately 670 billion base pairs. It seems that this is an infinitely long chain, so let's convert the size to our favorite meters. If all of our chromosomes (there are 46 of them, we don’t forget; 23 two copies each) turn and stretch into one line, we’ll get about a 2 meter chain. The DNA of one amoeba is enough to surround a football stadium. But what am I leading to? The nucleus in which the DNA is stored is not very large. We have an average diameter of 6 microns. Not very much if you want to roll a 2 meter thread, even if it is very thin. And you need not just push the thread into the core. It is necessary to turn in such a way that at any time it was possible to provide access to any of its parts. The task is complicated. And specialized proteins successfully cope with it. They create a series of spirals and loops that provide increasingly high levels of packaging and do not allow for DNA tangling into the Gordian knot. Let's talk about how it is packed. which provide increasingly higher levels of packaging and do not allow for DNA tangling into the Gordian knot. Let's talk about how it is packed. which provide increasingly higher levels of packaging and do not allow for DNA tangling into the Gordian knot. Let's talk about how it is packed.
I must say, it is packaged in very different ways. But if you throw away the exotic, then there are two ways. The first is characteristic of bacteria, the second for eukaryotes (or nuclear).
DNA packaging in bacteria
Let's start with our smaller brothers. Bacteria themselves do not have a very large genome, an average of 1 to 5 million base pairs. Their most characteristic difference from us is that they do not have a nucleus and the DNA is floating in the cell. Not quite floating, it is partially attached to the cell membrane and also collapsed, but not as much as ours.
The second. Bacterial DNA is often circular. So it is easier to copy (there are no ends that can be lost when copying and there is no need to invent mechanisms for saving ends). Usually such a ring is one, but some bacteria can have 2 or 3. There are also smaller rings (from a couple of thousands to a couple of hundreds of thousands of residues). They have a plasmid, and this is a different story altogether.
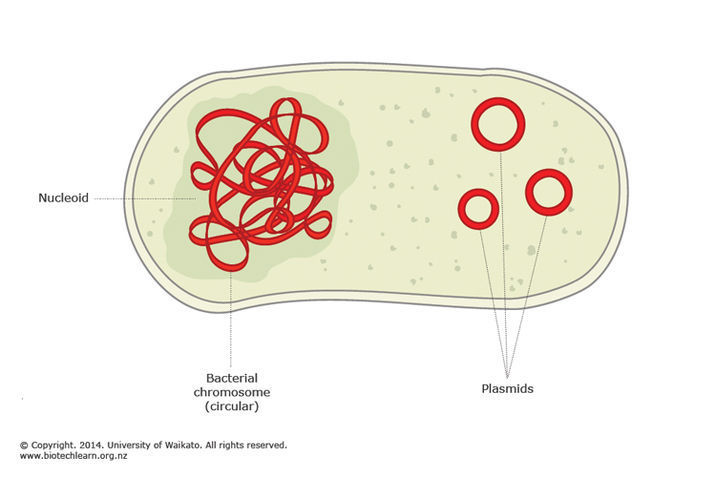
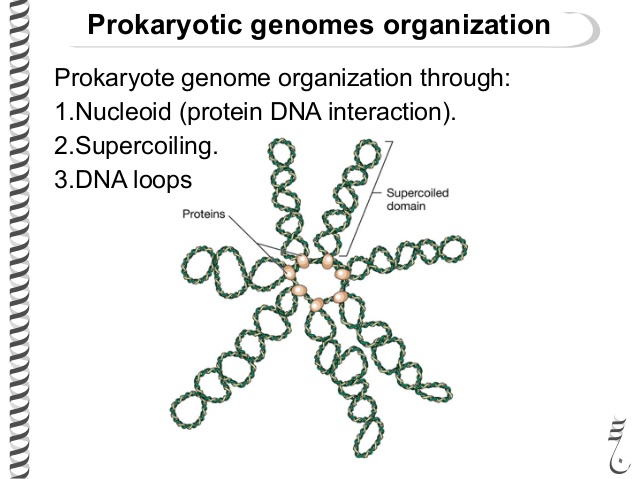
Let's go back to the DNA packaging. DNA is packed with histone proteins (there are also histone-like proteins). DNA is deoxyribonucleic acid. Acid . This means that it is negatively charged (due to residues of phosphoric acid). Therefore, the proteins that bind it are positively charged. In this way, they can bind to DNA. The DNA of bacteria together with its packaging proteins form a nucleoid, while DNA accounts for 80% of its mass. It looks like this. That is, the circular DNA is divided into domains of 40 thousand base pairs. Then twisting occurs. Twisting occurs within the domains as well, but its degree differs in different domains. On average, the degree of bacterial DNA packaging varies from a hundred to a thousand times.
There is still a cool video .
DNA packaging in eukaryotes
It's all much more interesting. Our DNA is well packed and hidden inside the nucleus. And it is much more efficiently packed than in bacteria. During mitosis (cell division), the size of chromosome 22 is 2 μm. If you unravel it and pull it out, it will already be 1.5 cm. That corresponds to a packing degree of 10,000 times. This is about the maximum degree of packing our DNA. During division, DNA needs to be packed as much as possible in order to effectively divide it between the daughter cells. In everyday life, the degree of compaction is about 500 times. Too packed DNA is difficult to read.
There are several levels of eukaryotic DNA packaging
The first is the nucleosome level. 8 histone proteins form a particle on which DNA is wound. Then another protein fixes it. It looks like this.
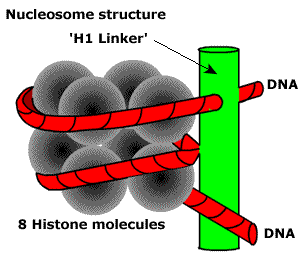

It turns out a kind of beads. Due to this, the packing density increases by 7-10 times. Next, nucleosomes are packaged into fibrils. A bit like a salt pans. Here the total degree of packing can reach 60 times.

The next stage of DNA compaction is associated with the formation of loop-like structures called chromomeres. Fibril is divided into sections of 10–80 thousand pairs of nitrogenous bases. Globules of nonhistone proteins are located in the breakdown sites. DNA binding proteins recognize globules of non-histone proteins and bring them together. Formed the mouth of the loop. The average loop length includes about 50 thousand bases. This structure is called interphase chromonema. And that is where our DNA is located most of the time. The level of packaging here reaches 500-1500 times.
If necessary, the cell can further compact the genetic material. The formation of larger loops of chromomeric fibrils. These loops in turn form new loops (loops in loops ... and this is not knitting). Which ultimately form the chromosome.
In general, the packaging process can be described something like this.

As a result, from the DNA strands we get, when dividing, supercoiled structures that can be seen under a microscope. We call them chromosomes.
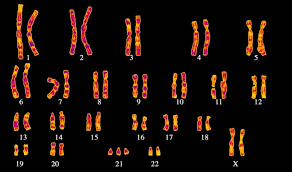
Actually the substance of chromosomes is called chromatin. And the degree of its packaging differs depending on the chromosome region. There is euchromatin and heterochromatin. Euchromatin is a rather unraveled region of chromatin, in which DNA is located at a chromomeric level (packing 500–1000 times). Here is the active reading of information. For example, if the cell is now actively synthesizing protein A, then the DNA region encoding it will be in the state of euchromatin, so that the enzymes that “read” the DNA could reach it. Heterochromatin also contains that part of the DNA that the cell does not really need right now. That is, the DNA is packed as tightly as possible so as not to get under your feet. Depending on the needs of the cell, some areas of chromatin can be partially untied, while others can be intertwined. Thus, regulation is also carried out (very rough approximation),
Actually, that's all for now. We discussed how the storage medium is stored. Let's make a short pause and in a couple of days we will talk about the coding of the information itself.
Only registered users can participate in the survey. Sign in , please.
