Molecular computer 3,000,000 centuries ago
They say that all inventions of mankind have been "noticed" by nature, or at least have natural analogues; for example, an airplane in nature has an analogue - a bird. Is there an analogue of a computer and computer programs?
This is a biological cell, which will be considered in this post from the point of view of a programmer, like a molecular computer . You will learn that many modern methods and algorithms for processing information did not appear in the 1980s, but many millions of years earlier.
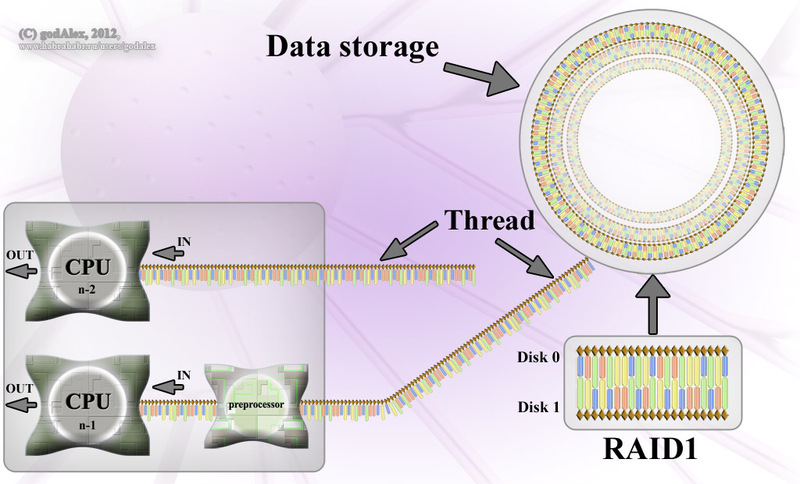
Data is stored on a molecular storage medium - on one molecule (DNA, in prokaryotes DNA in the form of a ring), and divided into clusters (genes). Data is stored as a 4-decimal code (physically represented as 4 nucleotides: adenine, thymine, cytosine, guanine). High fault tolerance and data integrity is carried out using RAID 1 technology (DNA is a double helix, nucleotides in both helices are connected in pairs A = T, C = G). Data integrity checks are carried out regularly (using DNA ligase protein), the damage found is restored, of course, with the duplicate remaining.
Replication of such a database is carried out by a special service (a protein complex called replicoma) by the method of physical separation of information carriers, immediately creating a full-fledged backup of one of the carriers, as well as checking the integrity of the information during the restoration and repairing the damage. Creating a backup copy for a replica is delayed to save resources (see DNA replication ). At the end of the replication process, both copies of the data contain backup copies that comply with the RAID 1 specification.
The medium also supports recording information ( reverse transcription ), but recording is rarely used in practice, usually difficult to destroy viruses ( retroviruses ).
Media Options:Memory capacity: scalable, replication speed:> 1 Gb / s (not accurate) , read / write speed: ??? Mb / s / ??? Mb / s
Since this is a molecular computer, the amount of RAM determines the number of freely accessible molecules that are not involved in data storage. When memory is allocated , “free” molecules are taken in RAM and arranged in threads, in a sequence encoding information.
When memory is freed, the filamentous molecules separate, replenishing the volume of free molecules.
At the beginning of reading data from the medium ( DNA transcription ) in the RAM, a search is made for the desired location on the medium ( binding sites ). Then, reading continues to the end of file sign ( presumably AAAA sequence). The resulting filamentous molecule (RNA or mRNA) is sent for processing.
Reading processes can take place in parallel.
The reading process is started by events (under the influence of special proteins ). When an event or group of events arrives, service information begins to be read from the desired piece of data on the medium (DNA), which may contain links to other sections that will be read for execution (initiation of transcription is a complex process depending on the DNA sequence and on the presence or absence of various protein factors).
There are many processors in the computer that execute code stored on whisker molecules (RNAs), so we can say that this type of computer supports multithreading, both literally and figuratively.
The word "stream", in a computer that was incorrectly once translated from English. Actually the translation literal thread will be - a thread!
The machine word is 3-bit, but 4-decimal. All commands of a fixed length are one machine word ( codon ), i.e. 64 kinds of teams.
Note: before it enters the processor, the stream (RNA) undergoes pre-processing ( splicing ), in which extra commands are removed.
The processor controls the molecular computer by creating proteins that do all the physical work: from building new structures and changing shape and movement to launching new programs (i.e., commands to read new RNAs).
Processor architecture: 64 command, execution speed: depends on the data bus (molecule delivery speed).
64
teams , of which: Code assembly information: 61 (however their actions are duplicated)
Controllers: 3 (conditional stop: UAG, UGA, mandatory UAA stop),
Reserved: approximately 41 can be used for other purposes if the processor is slightly changed (see . noncanonical codon ).
The code in the form of "assembler" was considered above, and now we will consider in the form of a super-high level language. It turned out that each gene is responsible for a specific function, or set of functions.
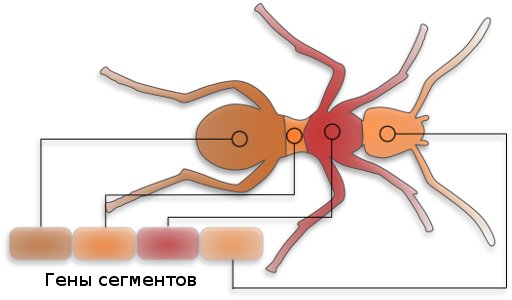
Multicellular organisms have genes that are responsible for constructing specific systems in living organisms, such as eyes, paws, etc. There are genes that are responsible for constructing segments: head, trunk, tail.
A change in the genes responsible for the internal structure of the eye will only affect its internal structure, and a change in the gene responsible for the location of the eye will cause the eyes to be in the body, but they will grow from other places. If you remove the gene responsible for the location of the eyes, then they will not grow.
It is noteworthy that these “blocks” (genes) are interchangeable in most living organisms. So, the scientists conducted an experiment: the gene responsible for the location of the eyes was removed from the Drosophila flies - new flies appeared eyeless. The gene responsible for the position of the eyes, which was taken from the mice, was inserted into the next generation of flies, and the flies had eyes, fly and not mouse.
Many genes seem to be the same for most living organisms, and most importantly, are interchangeable!
Scientists have already learned how to isolate genes from some living organisms, and insert them into others, getting new organisms. This process is called genetic engineering. As far as I know, now the functions of genes are detected by trial and error: they damage the gene, look at what has changed, or look at the “mutated” individual, and compare its genes with the reference one (something like cracking programs in the debugger).
When they understand what genes in many organisms are responsible for, when they learn not only to copy ready-made genes, but to create completely new ones, then they will not need to create any mechanical nanorobots - they will already be in the form of biologically programmed cells.
The first “DNA compiler” was made in 2010 by a group of scientists led by Dr. Craig Venter.
It was also successfully saved and then read abstract information in DNA (files about 600 kilobytes in size, and this is not a limitation) by specialists from the University of Cambridge. But you can read about it in other articles.
Sources and links:
Wikipedia: we look at all terms there. However, trust and verify - there are too many obviously inadequate numerical values, for example, the copy speed in this article was estimated with a very large error due to lack of information.
Documentary videos and films:
“Creating Synthetic Life” 2010 - editing DNA code on a computer, reading and writing to living cells, while there were surviving cells (a group of Dr. Craig Venter).
“BBC: Cell” 2009, “BBC: The Evolution of Life” - some episodes show the experience with Drosophila, which clearly show how genes are responsible for the location of body parts.
“Inner Life of a Cell” video for 8 minutes on youtub - without further ado the execution of a “navigation program” is shown ...
UPD : This article is licensed under Creative Commons CC-BY-NC since February 15, 2014.
This is a biological cell, which will be considered in this post from the point of view of a programmer, like a molecular computer . You will learn that many modern methods and algorithms for processing information did not appear in the 1980s, but many millions of years earlier.

Storage medium
Data is stored on a molecular storage medium - on one molecule (DNA, in prokaryotes DNA in the form of a ring), and divided into clusters (genes). Data is stored as a 4-decimal code (physically represented as 4 nucleotides: adenine, thymine, cytosine, guanine). High fault tolerance and data integrity is carried out using RAID 1 technology (DNA is a double helix, nucleotides in both helices are connected in pairs A = T, C = G). Data integrity checks are carried out regularly (using DNA ligase protein), the damage found is restored, of course, with the duplicate remaining.
Replication of such a database is carried out by a special service (a protein complex called replicoma) by the method of physical separation of information carriers, immediately creating a full-fledged backup of one of the carriers, as well as checking the integrity of the information during the restoration and repairing the damage. Creating a backup copy for a replica is delayed to save resources (see DNA replication ). At the end of the replication process, both copies of the data contain backup copies that comply with the RAID 1 specification.
The medium also supports recording information ( reverse transcription ), but recording is rarely used in practice, usually difficult to destroy viruses ( retroviruses ).
Media Options:Memory capacity: scalable, replication speed:> 1 Gb / s (not accurate) , read / write speed: ??? Mb / s / ??? Mb / s
RAM
Since this is a molecular computer, the amount of RAM determines the number of freely accessible molecules that are not involved in data storage. When memory is allocated , “free” molecules are taken in RAM and arranged in threads, in a sequence encoding information.
When memory is freed, the filamentous molecules separate, replenishing the volume of free molecules.
Reading data from a storage medium
At the beginning of reading data from the medium ( DNA transcription ) in the RAM, a search is made for the desired location on the medium ( binding sites ). Then, reading continues to the end of file sign ( presumably AAAA sequence). The resulting filamentous molecule (RNA or mRNA) is sent for processing.
Reading processes can take place in parallel.
Running event programs
The reading process is started by events (under the influence of special proteins ). When an event or group of events arrives, service information begins to be read from the desired piece of data on the medium (DNA), which may contain links to other sections that will be read for execution (initiation of transcription is a complex process depending on the DNA sequence and on the presence or absence of various protein factors).
CPU
There are many processors in the computer that execute code stored on whisker molecules (RNAs), so we can say that this type of computer supports multithreading, both literally and figuratively.
The word "stream", in a computer that was incorrectly once translated from English. Actually the translation literal thread will be - a thread!
The machine word is 3-bit, but 4-decimal. All commands of a fixed length are one machine word ( codon ), i.e. 64 kinds of teams.
Note: before it enters the processor, the stream (RNA) undergoes pre-processing ( splicing ), in which extra commands are removed.
The processor controls the molecular computer by creating proteins that do all the physical work: from building new structures and changing shape and movement to launching new programs (i.e., commands to read new RNAs).
Processor architecture: 64 command, execution speed: depends on the data bus (molecule delivery speed).
Assembler: which teams?
64
teams , of which: Code assembly information: 61 (however their actions are duplicated)
Controllers: 3 (conditional stop: UAG, UGA, mandatory UAA stop),
Reserved: approximately 41 can be used for other purposes if the processor is slightly changed (see . noncanonical codon ).
Modular Code Structure
The code in the form of "assembler" was considered above, and now we will consider in the form of a super-high level language. It turned out that each gene is responsible for a specific function, or set of functions.
Multicellular organisms have genes that are responsible for constructing specific systems in living organisms, such as eyes, paws, etc. There are genes that are responsible for constructing segments: head, trunk, tail.
A change in the genes responsible for the internal structure of the eye will only affect its internal structure, and a change in the gene responsible for the location of the eye will cause the eyes to be in the body, but they will grow from other places. If you remove the gene responsible for the location of the eyes, then they will not grow.
It is noteworthy that these “blocks” (genes) are interchangeable in most living organisms. So, the scientists conducted an experiment: the gene responsible for the location of the eyes was removed from the Drosophila flies - new flies appeared eyeless. The gene responsible for the position of the eyes, which was taken from the mice, was inserted into the next generation of flies, and the flies had eyes, fly and not mouse.
Many genes seem to be the same for most living organisms, and most importantly, are interchangeable!

What is genetic engineering?
Scientists have already learned how to isolate genes from some living organisms, and insert them into others, getting new organisms. This process is called genetic engineering. As far as I know, now the functions of genes are detected by trial and error: they damage the gene, look at what has changed, or look at the “mutated” individual, and compare its genes with the reference one (something like cracking programs in the debugger).
When they understand what genes in many organisms are responsible for, when they learn not only to copy ready-made genes, but to create completely new ones, then they will not need to create any mechanical nanorobots - they will already be in the form of biologically programmed cells.
Finally
The first “DNA compiler” was made in 2010 by a group of scientists led by Dr. Craig Venter.
It was also successfully saved and then read abstract information in DNA (files about 600 kilobytes in size, and this is not a limitation) by specialists from the University of Cambridge. But you can read about it in other articles.
Sources and links:
Wikipedia: we look at all terms there. However, trust and verify - there are too many obviously inadequate numerical values, for example, the copy speed in this article was estimated with a very large error due to lack of information.
Documentary videos and films:
“Creating Synthetic Life” 2010 - editing DNA code on a computer, reading and writing to living cells, while there were surviving cells (a group of Dr. Craig Venter).
“BBC: Cell” 2009, “BBC: The Evolution of Life” - some episodes show the experience with Drosophila, which clearly show how genes are responsible for the location of body parts.
“Inner Life of a Cell” video for 8 minutes on youtub - without further ado the execution of a “navigation program” is shown ...
UPD : This article is licensed under Creative Commons CC-BY-NC since February 15, 2014.
