Pandas Guide to Big Data Analysis
- Transfer
- Tutorial
When using the pandas library to analyze small datasets, the size of which does not exceed 100 megabytes, performance rarely becomes a problem. But when it comes to the study of data sets, the sizes of which can reach several gigabytes, performance problems can lead to a significant increase in the duration of data analysis and may even lead to the inability to conduct analysis due to lack of memory.
While tools like Spark can efficiently process large data sets (from hundreds of gigabytes to several terabytes), in order to fully utilize their capabilities, you usually need quite powerful and expensive hardware. And, in comparison with pandas, they do not differ in rich sets of tools for high-quality cleaning, research and data analysis. For medium-sized datasets, it's best to try using pandas more efficiently, rather than switching to other tools. In the material, the translation of which we publish today, we will talk about the peculiarities of working with memory when using pandas, and how, by simply selecting the appropriate data types stored in the columns of tabular data structures , reduce memory consumption by almost 90%.

We will work with data on Major League baseball games collected over 130 years and taken from Retrosheet .
Initially, this data was presented as 127 CSV files, but we combined them into one data set using csvkit and added, as the first row of the resulting table, a row with column names. If you want, you can download our version of this data and experiment with it, reading the article.
Let's start by importing a dataset and take a look at its first five lines. You can find them in this table, on the sheet
Below is information about the most important columns of the table with this data. If you want to read the explanations for all columns, here you can find a data dictionary for the entire data set.
In order to find out general information about the object
By default, pandas, for the sake of time saving, provides approximate information about the memory usage of the object
Here are the information we managed to get:
As it turned out, we have 171,907 rows and 161 columns. The pandas library automatically detected data types. There are 83 columns with numerical data and 78 columns with objects. Object columns are used to store string data, and in cases where the column contains data of different types.
Now, in order to better understand how you can optimize the memory usage of this object
Inside pandas, data columns are grouped into blocks with values of the same type. Here is an example of how pandas stores the first 12 columns of an object
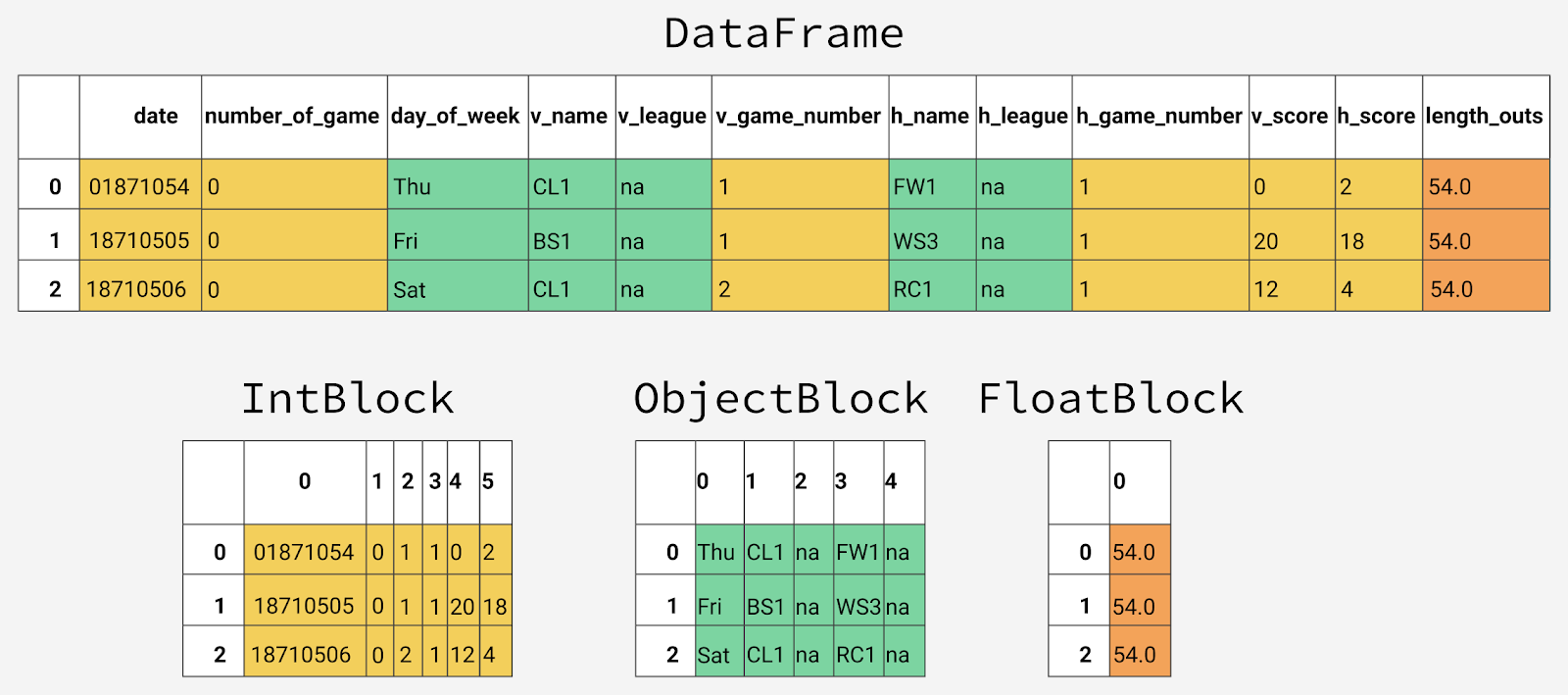
Internal representation of different types of data in pandas
You may notice that blocks do not store information about column names. This is due to the fact that the blocks are optimized for storing the values available in the cells of the object table
Each data type has a specialized class in the module
Since data of different types are stored separately, we examine the memory usage of different types of data. Let's start with the average memory usage for different types of data.
As a result, it turns out that the average indicators for memory usage for data of different types look like this:
This information allows us to understand that most of the memory is spent on 78 columns storing object values. We will talk about this later, but now let's think about whether we can improve memory usage with columns that store numerical data.
As we said, pandas represents numerical values as
Many data types in pandas have many subtypes that can use fewer bytes to represent each value. For example, a type
The type value
You can use the method to check the minimum and maximum values suitable for storage using each integer subtype
By executing this code, we get the following data:
Here you can pay attention to the difference between the types
The function
Here is the result of a study of memory consumption:
As a result, you can see a drop in memory usage from 7.9 to 1.5 megabytes, that is - we reduced memory consumption by more than 80%. The overall impact of this optimization on the original object
Let's do the same with columns containing floating point numbers.
The result is the following:
As a result, all columns that
Create a copy of the source object
Here's what we got: Although we significantly reduced memory consumption by columns storing numerical data, in general, over the whole object , memory consumption decreased by only 7%. The source of a much more serious improvement can be the optimization of storage of object types. Before we do this optimization, we’ll take a closer look at how strings are stored in pandas, and compare this with how numbers are stored here.
A type
This limitation leads to the fact that strings are not stored in contiguous fragments of memory; their representation in memory is fragmented. This leads to an increase in memory consumption and to a slowdown in the speed with string values. Each element in the column storing the object data type, in fact, is a pointer that contains the “address” at which the real value is located in memory.
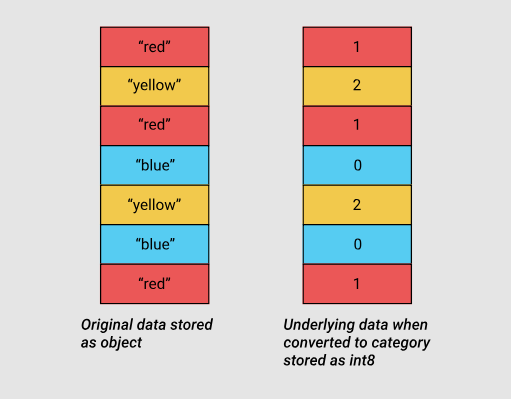
The diagram below is based on this. material that compares storing numerical data using NumPy data types and storing strings using Python's built-in data types.

Storing numeric and string data
Here you can recall that in one of the tables above it was shown that a variable amount of memory is used to store data of object types. Although each pointer occupies 1 byte of memory, each particular string value takes up the same amount of memory that would be used to store a single string in Python. In order to confirm this, we use the method
So, first we examine the usual lines:
Here, the data on memory usage looks like this: Now let's look at how the use of strings in the object looks :
Here we get the following:
Here you can see that the sizes of the lines stored in
Categorical variables appeared in pandas version 0.15. The corresponding type,,
Source data and categorical data using the int8 subtype
In order to understand exactly where we can use the categorical data to reduce memory consumption, we find out the number of unique values in the columns that store the values of object types:
What we have, you can find in this table, on the sheet
For example, in a column
Before we do full-scale optimization, let's select one column that stores object data, and at least
As already mentioned, this column contains only 7 unique values. To convert it to a categorical type, we use the method
Here's what we got:
As you can see, although the type of the column has changed, the data stored in it looks the same as before. Now let's look at what is happening inside the program.
In the following code, we use the attribute
We manage to find out the following:
Here you can see that each unique value is assigned an integer value, and that the column is now of type
Now let's compare the memory consumption before and after converting a column
Here's what happens: As you can see, at first 9.84 megabytes of memory were consumed, and after optimization it was only 0.16 megabytes, which means a 98% improvement in this indicator. Please note that working with this column probably demonstrates one of the most profitable optimization scenarios when only 7 unique values are used in a column containing approximately 172,000 elements. Although the idea of converting all columns to this data type looks attractive, before doing this, consider the negative side effects of such a conversion. So, the most serious minus of this transformation is the impossibility of performing arithmetic operations on categorical data. This also applies to ordinary arithmetic operations, and the use of methods like and
We should limit the use of the type
Let's create a loop that iterates over all the columns that store the type data
Now compare what happened after optimization with what happened before:
We get the following:
In our case, all processed columns were converted to type
As you can see, the amount of memory needed to work with columns storing type data
Here's what we got:
The result is impressive. But we can still improve something. As shown above, in our table there are data of the type
In terms of memory usage, the following is obtained here:
Here is a summary of the data:
You may recall that the source data was presented in integer form and was already optimized using the type
The conversion is performed using a function
The result is the following:
Data now looks like this:
So far, we have explored ways to reduce memory consumption by an existing object
Fortunately, the optimal data types for individual columns can be specified even before the actual data loading. Pandas.read_csv () functionhas several options to do this. So, the parameter
In order to use this technique, we will save the final types of all columns in a dictionary with keys represented by column names. But first, remove the column with the date of the game, since it needs to be processed separately.
Now we will be able to use this dictionary along with several parameters related to the data on the dates of the games during the data download.
The corresponding code is pretty compact:
As a result, the amount of memory used looks like this: The
data now looks like the one shown in the sheet
Externally, the tables shown on the sheets
Now, after we have optimized the data, we can do their analysis. Take a look at the distribution of gaming days.

Days during which games were held
As you can see, until the 1920s, games were rarely held on Sundays, after which, for about 50 years, games on that day were gradually held more often.
In addition, you can see that the distribution of the days of the week in which games have been played for the past 50 years is practically unchanged.
Now let's take a look at how the duration of games has changed over time.
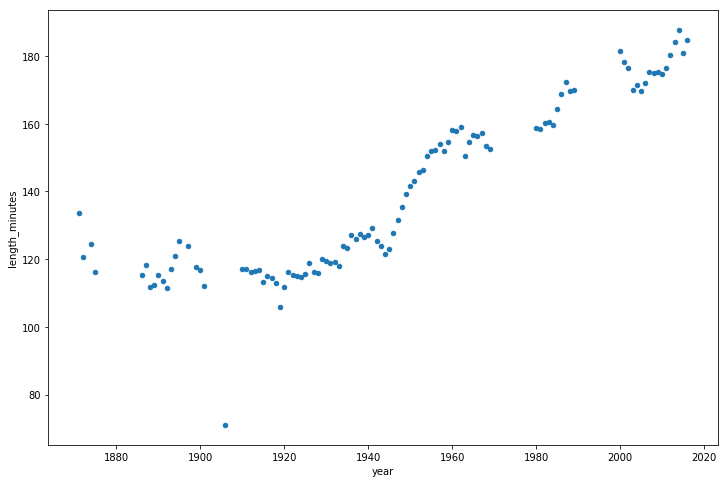
Duration of games
It seems that from the 1940s to the present, matches are becoming longer.
In this article, we discussed the features of storing data of various types in pandas, after which we used the knowledge we obtained to reduce the amount of memory needed to store an object
This is not to say that the optimization of any data set can lead to equally impressive results, but, especially considering the possibility of performing the optimization at the data loading stage, we can say that it is useful for anyone who is involved in data analysis using pandas to own working methods that we discussed here.
Dear readers! To translate this article, our reader recommended us eugene_bb . If you know of any interesting materials that are worth translating, tell us about them.

While tools like Spark can efficiently process large data sets (from hundreds of gigabytes to several terabytes), in order to fully utilize their capabilities, you usually need quite powerful and expensive hardware. And, in comparison with pandas, they do not differ in rich sets of tools for high-quality cleaning, research and data analysis. For medium-sized datasets, it's best to try using pandas more efficiently, rather than switching to other tools. In the material, the translation of which we publish today, we will talk about the peculiarities of working with memory when using pandas, and how, by simply selecting the appropriate data types stored in the columns of tabular data structures , reduce memory consumption by almost 90%.

DataFrameWorking with data on baseball games
We will work with data on Major League baseball games collected over 130 years and taken from Retrosheet .
Initially, this data was presented as 127 CSV files, but we combined them into one data set using csvkit and added, as the first row of the resulting table, a row with column names. If you want, you can download our version of this data and experiment with it, reading the article.
Let's start by importing a dataset and take a look at its first five lines. You can find them in this table, on the sheet
Фрагмент исходного набора данных.import pandas as pd
gl = pd.read_csv('game_logs.csv')
gl.head()Below is information about the most important columns of the table with this data. If you want to read the explanations for all columns, here you can find a data dictionary for the entire data set.
date- Date of the game.v_name- Name of the guest team.v_league- League guest team.h_name- The name of the home team.h_league- The home team league.v_score- Points of the guest team.h_score- Points of the home team.v_line_score- A summary of the points of the guest team, for example -010000(10)00.h_line_score- A summary of the points of the home team, for example -010000(10)0X.park_id- The identifier of the field on which the game was played.attendance- The number of viewers.
In order to find out general information about the object
DataFrame, you can use the DataFrame.info () method . Thanks to this method, you can learn about the size of an object, about data types, and about memory usage. By default, pandas, for the sake of time saving, provides approximate information about the memory usage of the object
DataFrame. We are interested in accurate information, so we will set the parameter memory_usageto a value 'deep'.gl.info(memory_usage='deep')Here are the information we managed to get:
RangeIndex: 171907 entries, 0 to 171906
Columns: 161 entries, date to acquisition_info
dtypes: float64(77), int64(6), object(78)
memory usage: 861.6 MB As it turned out, we have 171,907 rows and 161 columns. The pandas library automatically detected data types. There are 83 columns with numerical data and 78 columns with objects. Object columns are used to store string data, and in cases where the column contains data of different types.
Now, in order to better understand how you can optimize the memory usage of this object
DataFrame, let's talk about how pandas stores data in memory.Internal View of a DataFrame
Inside pandas, data columns are grouped into blocks with values of the same type. Here is an example of how pandas stores the first 12 columns of an object
DataFrame.
Internal representation of different types of data in pandas
You may notice that blocks do not store information about column names. This is due to the fact that the blocks are optimized for storing the values available in the cells of the object table
DataFrame. The class is responsible for storing information about the correspondence between the row and column indexes of the data set and what is stored in blocks of the same type of data BlockManager. It plays the role of an API that provides access to basic data. When we read, edit, or delete values, the class DataFrameinteracts with the class BlockManagerto convert our queries into function and method calls. Each data type has a specialized class in the module
pandas.core.internals. For example, pandas uses the classObjectBlockfor representing blocks containing string columns, and a class FloatBlockfor representing blocks containing columns holding floating-point numbers. For blocks representing numeric values that look like integers or floating point numbers, pandas combines the columns and stores them as a ndarrayNumPy library data structure . This data structure is built on the basis of array C, the values are stored in a continuous block of memory. Thanks to this data storage scheme, access to data fragments is very fast. Since data of different types are stored separately, we examine the memory usage of different types of data. Let's start with the average memory usage for different types of data.
for dtype in ['float','int','object']:
selected_dtype = gl.select_dtypes(include=[dtype])
mean_usage_b = selected_dtype.memory_usage(deep=True).mean()
mean_usage_mb = mean_usage_b / 1024 ** 2
print("Average memory usage for {} columns: {:03.2f} MB".format(dtype,mean_usage_mb))As a result, it turns out that the average indicators for memory usage for data of different types look like this:
Average memory usage for float columns: 1.29 MB
Average memory usage for int columns: 1.12 MB
Average memory usage for object columns: 9.53 MBThis information allows us to understand that most of the memory is spent on 78 columns storing object values. We will talk about this later, but now let's think about whether we can improve memory usage with columns that store numerical data.
Subtypes
As we said, pandas represents numerical values as
ndarrayNumPy data structures and stores them in contiguous blocks of memory. This data storage model allows you to save memory and quickly access values. Since pandas represents each value of the same type using the same number of bytes, and the structures ndarraystore information about the number of values, pandas can quickly and accurately display information about the amount of memory consumed by columns storing numerical values. Many data types in pandas have many subtypes that can use fewer bytes to represent each value. For example, a type
floathas subtypes float16, float32andfloat64. The number in the type name indicates the number of bits that the subtype uses to represent the values. For example, in the subtypes just listed, 2, 4, 8, and 16 bytes are used, respectively. The following table shows the subtypes of the most commonly used data types in pandas.| Memory usage, bytes | Floating point number | Integer | Unsigned integer | date and time | Boolean value | An object |
| 1 | int8 | uint8 | bool | |||
| 2 | float16 | int16 | uint16 | |||
| 4 | float32 | int32 | uint32 | |||
| 8 | float64 | int64 | uint64 | datetime64 | ||
| Variable memory capacity | object |
The type value
int8uses 1 byte (8 bits) to store the number and can represent 256 binary values (2 to the 8th power). This means that this subtype can be used to store values in the range from -128 to 127 (including 0). You can use the method to check the minimum and maximum values suitable for storage using each integer subtype
numpy.iinfo(). Consider an example:import numpy as np
int_types = ["uint8", "int8", "int16"]
for it in int_types:
print(np.iinfo(it))By executing this code, we get the following data:
Machine parameters for uint8
---------------------------------------------------------------
min = 0
max = 255
---------------------------------------------------------------
Machine parameters for int8
---------------------------------------------------------------
min = -128
max = 127
---------------------------------------------------------------
Machine parameters for int16
---------------------------------------------------------------
min = -32768
max = 32767
---------------------------------------------------------------Here you can pay attention to the difference between the types
uint(unsigned integer) and int(signed integer). Both types have the same capacity, but when storing only positive values in columns, unsigned types allow more efficient use of memory.Optimization of storage of numerical data using subtypes
The function
pd.to_numeric()can be used for the downward conversion of numeric types. To select integer columns, we use the method DataFrame.select_dtypes(), then we optimize them and compare the memory usage before and after optimization.# Мы будем часто выяснять то, сколько памяти используется,
# поэтому создадим функцию, которая поможет нам сэкономить немного времени.
def mem_usage(pandas_obj):
if isinstance(pandas_obj,pd.DataFrame):
usage_b = pandas_obj.memory_usage(deep=True).sum()
else: # исходим из предположения о том, что если это не DataFrame, то это Series
usage_b = pandas_obj.memory_usage(deep=True)
usage_mb = usage_b / 1024 ** 2 # преобразуем байты в мегабайты
return "{:03.2f} MB".format(usage_mb)
gl_int = gl.select_dtypes(include=['int'])
converted_int = gl_int.apply(pd.to_numeric,downcast='unsigned')
print(mem_usage(gl_int))
print(mem_usage(converted_int))
compare_ints = pd.concat([gl_int.dtypes,converted_int.dtypes],axis=1)
compare_ints.columns = ['before','after']
compare_ints.apply(pd.Series.value_counts)Here is the result of a study of memory consumption:
7.87 MB
1.48 MB| Before | After | |
| uint8 | NaN | 5.0 |
| uint32 | NaN | 1.0 |
| int64 | 6.0 | NaN |
As a result, you can see a drop in memory usage from 7.9 to 1.5 megabytes, that is - we reduced memory consumption by more than 80%. The overall impact of this optimization on the original object
DataFrame, however, is not particularly strong, since it has very few integer columns. Let's do the same with columns containing floating point numbers.
gl_float = gl.select_dtypes(include=['float'])
converted_float = gl_float.apply(pd.to_numeric,downcast='float')
print(mem_usage(gl_float))
print(mem_usage(converted_float))
compare_floats = pd.concat([gl_float.dtypes,converted_float.dtypes],axis=1)
compare_floats.columns = ['before','after']
compare_floats.apply(pd.Series.value_counts)The result is the following:
100.99 MB
50.49 MB| Before | After | |
| float32 | NaN | 77.0 |
| float64 | 77.0 | NaN |
As a result, all columns that
float64store floating-point numbers with a data type now store type numbers float32, which gives us a 50% reduction in memory usage. Create a copy of the source object
DataFrame, use these optimized numeric columns instead of those that were originally present in it, and look at the overall memory usage after optimization.optimized_gl = gl.copy()
optimized_gl[converted_int.columns] = converted_int
optimized_gl[converted_float.columns] = converted_float
print(mem_usage(gl))
print(mem_usage(optimized_gl))Here's what we got: Although we significantly reduced memory consumption by columns storing numerical data, in general, over the whole object , memory consumption decreased by only 7%. The source of a much more serious improvement can be the optimization of storage of object types. Before we do this optimization, we’ll take a closer look at how strings are stored in pandas, and compare this with how numbers are stored here.
861.57 MB
804.69 MBDataFrameComparison of mechanisms for storing numbers and strings
A type
objectrepresents values using Python string objects. This is partly because NumPy does not support the representation of missing string values. Since Python is a high-level interpreted language, it does not provide the programmer with tools for fine-tuning how data is stored in memory. This limitation leads to the fact that strings are not stored in contiguous fragments of memory; their representation in memory is fragmented. This leads to an increase in memory consumption and to a slowdown in the speed with string values. Each element in the column storing the object data type, in fact, is a pointer that contains the “address” at which the real value is located in memory.
The diagram below is based on this. material that compares storing numerical data using NumPy data types and storing strings using Python's built-in data types.

Storing numeric and string data
Here you can recall that in one of the tables above it was shown that a variable amount of memory is used to store data of object types. Although each pointer occupies 1 byte of memory, each particular string value takes up the same amount of memory that would be used to store a single string in Python. In order to confirm this, we use the method
sys.getsizeof(). First, take a look at the individual lines, and then at the Seriespandas object that stores the string data. So, first we examine the usual lines:
from sys import getsizeof
s1 = 'working out'
s2 = 'memory usage for'
s3 = 'strings in python is fun!'
s4 = 'strings in python is fun!'
for s in [s1, s2, s3, s4]:
print(getsizeof(s))Here, the data on memory usage looks like this: Now let's look at how the use of strings in the object looks :
60
65
74
74Seriesobj_series = pd.Series(['working out',
'memory usage for',
'strings in python is fun!',
'strings in python is fun!'])
obj_series.apply(getsizeof)Here we get the following:
0 60
1 65
2 74
3 74
dtype: int64Here you can see that the sizes of the lines stored in
Seriespandas objects are similar to their sizes when working with them in Python and when representing them as independent entities.Optimization of storage of data of object types using categorical variables
Categorical variables appeared in pandas version 0.15. The corresponding type,,
categoryuses integer values in its internal mechanisms, instead of the original values stored in the table columns. Pandas uses a separate dictionary that sets the correspondence of integer and initial values. This approach is useful when the columns contain values from a limited set. When the data stored in a column is converted to a type category, pandas uses a subtype intthat allows it to manage memory most efficiently and is able to represent all the unique values found in the column.
Source data and categorical data using the int8 subtype
In order to understand exactly where we can use the categorical data to reduce memory consumption, we find out the number of unique values in the columns that store the values of object types:
gl_obj = gl.select_dtypes(include=['object']).copy()
gl_obj.describe()What we have, you can find in this table, on the sheet
Количество уникальных значений в столбцах. For example, in a column
day_of_weekrepresenting the day of the week the game was played, there are 171907 values. Among them, only 7 are unique. On the whole, a single glance at this report is enough to understand that quite a few unique values are used in many columns to represent the data of approximately 172,000 games. Before we do full-scale optimization, let's select one column that stores object data, and at least
day_of_weeksee what happens inside the program when it is converted to a categorical type.As already mentioned, this column contains only 7 unique values. To convert it to a categorical type, we use the method
.astype().dow = gl_obj.day_of_week
print(dow.head())
dow_cat = dow.astype('category')
print(dow_cat.head())Here's what we got:
0 Thu
1 Fri
2 Sat
3 Mon
4 Tue
Name: day_of_week, dtype: object
0 Thu
1 Fri
2 Sat
3 Mon
4 Tue
Name: day_of_week, dtype: category
Categories (7, object): [Fri, Mon, Sat, Sun, Thu, Tue, Wed]As you can see, although the type of the column has changed, the data stored in it looks the same as before. Now let's look at what is happening inside the program.
In the following code, we use the attribute
Series.cat.codesto find out which integer values the type categoryuses to represent each day of the week:dow_cat.head().cat.codesWe manage to find out the following:
0 4
1 0
2 2
3 1
4 5
dtype: int8Here you can see that each unique value is assigned an integer value, and that the column is now of type
int8. There are no missing values, but if that were the case, -1 would be used to indicate such values. Now let's compare the memory consumption before and after converting a column
day_of_weekto a type category.print(mem_usage(dow))
print(mem_usage(dow_cat))Here's what happens: As you can see, at first 9.84 megabytes of memory were consumed, and after optimization it was only 0.16 megabytes, which means a 98% improvement in this indicator. Please note that working with this column probably demonstrates one of the most profitable optimization scenarios when only 7 unique values are used in a column containing approximately 172,000 elements. Although the idea of converting all columns to this data type looks attractive, before doing this, consider the negative side effects of such a conversion. So, the most serious minus of this transformation is the impossibility of performing arithmetic operations on categorical data. This also applies to ordinary arithmetic operations, and the use of methods like and
9.84 MB
0.16 MBSeries.min()Series.max()without first converting the data to a real numeric type. We should limit the use of the type
categoryto mainly columns storing type data objectin which less than 50% of the values are unique. If all values in a column are unique, then using a type categorywill increase the level of memory usage. This is due to the fact that in memory you have to store, in addition to the numeric category codes, the original string values. Details on type restrictions categorycan be found in the pandas documentation . Let's create a loop that iterates over all the columns that store the type data
object, finds out if the number of unique values in the columns does not exceed 50%, and if so, converts them into a type category.converted_obj = pd.DataFrame()
for col in gl_obj.columns:
num_unique_values = len(gl_obj[col].unique())
num_total_values = len(gl_obj[col])
if num_unique_values / num_total_values < 0.5:
converted_obj.loc[:,col] = gl_obj[col].astype('category')
else:
converted_obj.loc[:,col] = gl_obj[col]Now compare what happened after optimization with what happened before:
print(mem_usage(gl_obj))
print(mem_usage(converted_obj))
compare_obj = pd.concat([gl_obj.dtypes,converted_obj.dtypes],axis=1)
compare_obj.columns = ['before','after']
compare_obj.apply(pd.Series.value_counts)We get the following:
752.72 MB
51.67 MB| Before | After | |
| object | 78.0 | NaN |
| category | NaN | 78.0 |
In our case, all processed columns were converted to type
category, however, we cannot say that the same thing will happen when processing any data set, therefore, when processing your data using this technique, do not forget about comparisons of what happened before the optimization, what happened after its implementation. As you can see, the amount of memory needed to work with columns storing type data
objecthas decreased from 752 megabytes to 52 megabytes, that is, 93%. Now let's look at how we were able to optimize memory consumption across the entire data set. Let us analyze what level of memory use we have reached, if we compare what happened with the initial indicator of 891 megabytes.optimized_gl[converted_obj.columns] = converted_obj
mem_usage(optimized_gl)Here's what we got:
'103.64 MB'The result is impressive. But we can still improve something. As shown above, in our table there are data of the type
datetimewhose storage column can be used as the first column of the data set.date = optimized_gl.date
print(mem_usage(date))
date.head()In terms of memory usage, the following is obtained here:
0.66 MBHere is a summary of the data:
0 18710504
1 18710505
2 18710506
3 18710508
4 18710509
Name: date, dtype: uint32You may recall that the source data was presented in integer form and was already optimized using the type
uint32. Because of this, converting this data to a type datetimewill double the memory consumption, since this type uses 64 bits to store data. However, in the conversion of data to type datetime, it still makes sense, since this will allow us to more easily perform time series analysis. The conversion is performed using a function
to_datetime()whose parameter formatindicates that the data is stored in a format YYYY-MM-DD.optimized_gl['date'] = pd.to_datetime(date,format='%Y%m%d')
print(mem_usage(optimized_gl))
optimized_gl.date.head()The result is the following:
104.29 MBData now looks like this:
0 1871-05-04
1 1871-05-05
2 1871-05-06
3 1871-05-08
4 1871-05-09
Name: date, dtype: datetime64[ns]Type selection when loading data
So far, we have explored ways to reduce memory consumption by an existing object
DataFrame. We first read the data in its original form, then, step by step, engaged in their optimization, comparing what happened with what happened. This allowed us to properly understand what can be expected from certain optimizations. As already mentioned, often there may simply not be enough memory to represent all the values included in a certain data set. In this regard, the question arises of how to apply memory saving techniques in the event that it is impossible even to create an object DataFramethat is supposed to be optimized. Fortunately, the optimal data types for individual columns can be specified even before the actual data loading. Pandas.read_csv () functionhas several options to do this. So, the parameter
dtypeaccepts a dictionary in which, in the form of keys, string names of columns are present, and in the form of values are NumPy types. In order to use this technique, we will save the final types of all columns in a dictionary with keys represented by column names. But first, remove the column with the date of the game, since it needs to be processed separately.
dtypes = optimized_gl.drop('date',axis=1).dtypes
dtypes_col = dtypes.index
dtypes_type = [i.name for i in dtypes.values]
column_types = dict(zip(dtypes_col, dtypes_type))
# вместо вывода всех 161 элементов, мы
# возьмём 10 пар ключ/значение из словаря
# и аккуратно их выведем
preview = first2pairs = {key:value for key,value in list(column_types.items())[:10]}
import pprint
pp = pp = pprint.PrettyPrinter(indent=4)
pp.pprint(preview)
Вот что у нас получится:
{ 'acquisition_info': 'category',
'h_caught_stealing': 'float32',
'h_player_1_name': 'category',
'h_player_9_name': 'category',
'v_assists': 'float32',
'v_first_catcher_interference': 'float32',
'v_grounded_into_double': 'float32',
'v_player_1_id': 'category',
'v_player_3_id': 'category',
'v_player_5_id': 'category'}Now we will be able to use this dictionary along with several parameters related to the data on the dates of the games during the data download.
The corresponding code is pretty compact:
read_and_optimized = pd.read_csv('game_logs.csv',dtype=column_types,parse_dates=['date'],infer_datetime_format=True)
print(mem_usage(read_and_optimized))
read_and_optimized.head()As a result, the amount of memory used looks like this: The
104.28 MBdata now looks like the one shown in the sheet
Фрагмент оптимизированного набора данныхin this table. Externally, the tables shown on the sheets
Фрагмент оптимизированного набора данныхand Фрагмент исходного набора данных, with the exception of the column with the dates, look the same, but this only applies to their appearance. Thanks to the optimization of memory usage in pandas, we were able to reduce memory consumption from 861.6 MB to 104.28 MB, having an impressive result of saving 88% of memory.Analysis of baseball matches
Now, after we have optimized the data, we can do their analysis. Take a look at the distribution of gaming days.
optimized_gl['year'] = optimized_gl.date.dt.year
games_per_day = optimized_gl.pivot_table(index='year',columns='day_of_week',values='date',aggfunc=len)
games_per_day = games_per_day.divide(games_per_day.sum(axis=1),axis=0)
ax = games_per_day.plot(kind='area',stacked='true')
ax.legend(loc='upper right')
ax.set_ylim(0,1)
plt.show()
Days during which games were held
As you can see, until the 1920s, games were rarely held on Sundays, after which, for about 50 years, games on that day were gradually held more often.
In addition, you can see that the distribution of the days of the week in which games have been played for the past 50 years is practically unchanged.
Now let's take a look at how the duration of games has changed over time.
game_lengths = optimized_gl.pivot_table(index='year', values='length_minutes')
game_lengths.reset_index().plot.scatter('year','length_minutes')
plt.show()
Duration of games
It seems that from the 1940s to the present, matches are becoming longer.
Summary
In this article, we discussed the features of storing data of various types in pandas, after which we used the knowledge we obtained to reduce the amount of memory needed to store an object
DataFrameby almost 90%. To do this, we applied two simple techniques:- We performed a downward conversion of the types of numerical data stored in columns, choosing more efficient types in terms of memory usage.
- We converted string data to a categorical data type.
This is not to say that the optimization of any data set can lead to equally impressive results, but, especially considering the possibility of performing the optimization at the data loading stage, we can say that it is useful for anyone who is involved in data analysis using pandas to own working methods that we discussed here.
Dear readers! To translate this article, our reader recommended us eugene_bb . If you know of any interesting materials that are worth translating, tell us about them.

