DNA Mechanisms for storing and processing information. Part II

Hi Habr! Today we will continue the last story about DNA. In it, we talked about how much it is, how DNA is stored and why it is so important . Today we begin with a historical background and finish with the basics of coding information in DNA.
Story
By itself, DNA was isolated back in 1869 by Johann Friedrich Miescher from leukocytes, which he received from pus. Leukocytes are white blood cells that perform a protective function. There are a lot of pus in them, because they tend to damaged tissues, where they are “eaten” by bacterial cells. He isolated a substance that contains nitrogen and phosphorus. Initially, it was named nuclein, however, when it was discovered acidic properties, the name was changed to nucleic acid. The biological function of the newly discovered substance was unclear, and for a long time it was considered that phosphorus was stored in it. Even at the beginning of the 20th century, many biologists believed that DNA had nothing to do with information transfer, since the structure of the molecule, as it seemed, was too monotonous and could not encode so much information.
By 1901, Albrecht Kossel identified and described five nitrogenous bases that make up DNA and RNA. And a little later, Peter Leven found that the carbohydrate component of nucleic acids are deoxyribose and ribose. Nucleic acids, which include ribose, became known as ribonucleic acids or, in short, RNA, and those that contained deoxyribose, deoxyribonucleic acids, or DNA.
Now, the question arose how the individual links are interconnected. To do this, the DNA chain had to be broken down and look at what happens after the destruction. For this polymer DNA was subjected to hydrolysis. However, Leven changed the method of hydrolysis. Now instead of boiling for hours in an acidic medium, he used enzymes. This time, it was possible to isolate not only individual adenine, guanine, thymine, cytosine, deoxyribose and phosphoric acid from hydrolysates, but also larger fragments, for example, compounds of nitrogenous bases with carbohydrate or carbohydrate with phosphoric acid. At the same time , compounds consisting of two nitrogenous bases or compounds of the base-phosphoric acid type were not found in nucleic acid hydrolysates.. That is, it became clear that phosphoric acid combines with sugar, and he, in turn, with a nitrogenous base. Compounds of nitrogenous bases with carbohydrate were suggested to be called nucleosides, and phosphoric esters of nucleosides were called nucleotides.
As a result of these works, Leven concluded that nucleic acids are polymers. Nucleotides are used as monomers. The content of each of the four nucleotides in DNA, or RNA, according to chemical analysis of that time, seemed to Leven equal. Therefore, Leven proposed the following theory of the structure of nucleic acids: they are polymers whose monomers are blocks of four nucleotides connected in series.
The theory of the tetranucleotide structure at that time looked quite reasonable, entering into all the textbooks before the war. However, the question of DNA function remained unclear. To clarify this issue took almost half a century.
A period began during which biologists accumulated information on the distribution of nucleic acids in various types of animal and plant tissues, in bacteria and viruses, in some unicellular organisms.
At that time, the scientific community seriously believed that it was proteins that were responsible for storing genetic information. The traditional understanding of the primary role of proteins in the life process did not even allow thinking that such an important substance as the substance of heredity could be anything other than protein. Proteins were extremely diverse in their structure, which then could not be said about nucleic acids. The famous Soviet geneticist-cytologist N. K. Koltsov calculated that by varying the sequence of 20 amino acids that make up a protein molecule, trillions of proteins unlike each other can be created.
If we wanted to print in the most simplified form, as the logarithmic tables are printed, this trillion molecules and provided all the existing printing presses of the world to perform this plan, releasing 50,000 volumes of 100 printed pages per year, then so much time had passed how many have passed since the Archean period d of our days.
Indeed a lot ... 20 to 20th ... But the sequences are much longer than 20 amino acids.
But as R. R. Kizel writes about this, one of the most erudite biochemists of that time.
From the views on the role of nucleic acid just cited ... it follows that it does not belong to the structure of genes and it follows that the genes are composed of some other material. We still do not reliably know this material, despite the fact that in most cases it is directly called protein.The first success came from microbiology. In 1944, the results of the experiments of Avery and his colleagues (USA) on the transformation of bacteria were published. A few words about transformation.
The transformation itself was discovered in 1928 by the microbiologist Griffiths.
Griffith worked with pneumococcal cultures (Streptococcus pneumoniae) of the causative agent of a form of pneumonia. Some strains of this bacterium are virulent, causing pneumonia. Their cells are covered with a polysaccharide capsule that protects the bacteria from the action of the immune system. In culture, such bacteria form large smooth colonies of regular spherical shape. Because of this, they were called S – strains (from the English smooth - smooth).
There are various virulent pneumococcal strains, they differ in antibodies that are produced in the body when bacteria enter it. They are called IS, IIS, IIIS, etc. From time to time, some cells of the virulent S strains mutate, losing the ability to synthesize the polysaccharide membrane, and become avirulent. In culture, they form small, rough colonies of irregular shape, because of this they are called R – strains (from English rough - rough). Sometimes there are reverse mutations that restore the ability to synthesize a polysaccharide shell, but only in groups of the corresponding strains:
IIS - IIR
IIIS - IIIR
This suggests that avirulent R-strains always correspond to the parent virulent S-strain.
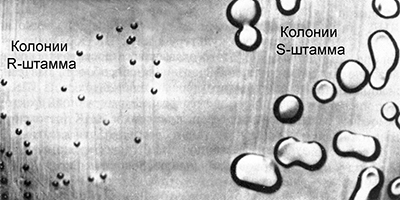
Griffith introduced to different groups of laboratory mice a virulent and avirulent strain of pneumococcus. In the first control group, injection of a virulent IIIS strain resulted in the death of animals. Animals of the second control group after injection of the avirulent IIR strain remained alive. After that, Griffith heated a solution with a culture of a virulent strain IIIS at a temperature of 60 ° C, which led to the death of bacteria. Killed by heating bacteria, he introduced the third group of experimental mice. The animals remained alive, which in principle was expected. However, this is not all. He introduced parts of the surviving mice bacteria of the avirulent IIR strain.
It seemed that this could not lead to any terrible consequences for mice. However, contrary to expectations, the animals died. When bacteria were isolated from their bodies and cultured, it turned out that they belong to the virulent strain IIIS.

The fact that the death-causing cells of the mice synthesized a type III rather than II polysaccharide membrane showed that they could not result from a reverse IIR-IIS mutation. From this Griffith made a very important conclusion. Avirulent bacteria of the IIR strain can be transformed into virulent bacteria somehow interacting with the heat-killed bacteria of the IIIS strain that still remained in the body of the mice. In other words, the avirulent bacteria of the IIR strain receive from the dead bacteria of the IIIS strain some factor that turns them into virulence. However, what is this factor, Griffith did not know.
Actually this phenomenon was called bacterial transformation. It is a unidirectional transfer of inherited traits from one bacterial cell to another.
Now back to the experiments of Avery. The scheme of their experiments is somewhat similar to the experiments of Griffiths. Avery and employees set themselves the task of finding out the chemical nature of the transforming agent. They destroyed the suspension of pneumococci and removed proteins, capsular polysaccharide and RNA from the extract, however, the transforming activity of the extract remained. The transforming activity of the drug was not lost when it was treated with crystalline trypsin or chymotrypsin (destroying proteins), ribonuclease (destroying RNA). It was clear that the drug was neither protein nor RNA. However, the transforming activity of the drug was completely lost when it was treated with deoxyribonuclease (destructive DNA), and insignificant amounts of the enzyme caused complete inactivation of the drug. Thus, it was established that the transforming factor in bacteria is pure DNA. This conclusion was a significant discovery, and Avery was perfectly aware of this. He wrote that this is exactly what genetics had long dreamed of, namely the substance of the gene. Seems here it is proof. But belief in protein, like the substance of heredity, was too strong. Some believed that the insignificant impurities of protein that remained in the preparation could also cause transformation.
New evidence of the direct genetic role of DNA was the experiments of virologists Hershey and Chase. They worked with bacteriophage T2 (Bacteriophages - bacteria viruses) that infect Escherichia coli (E. coli).
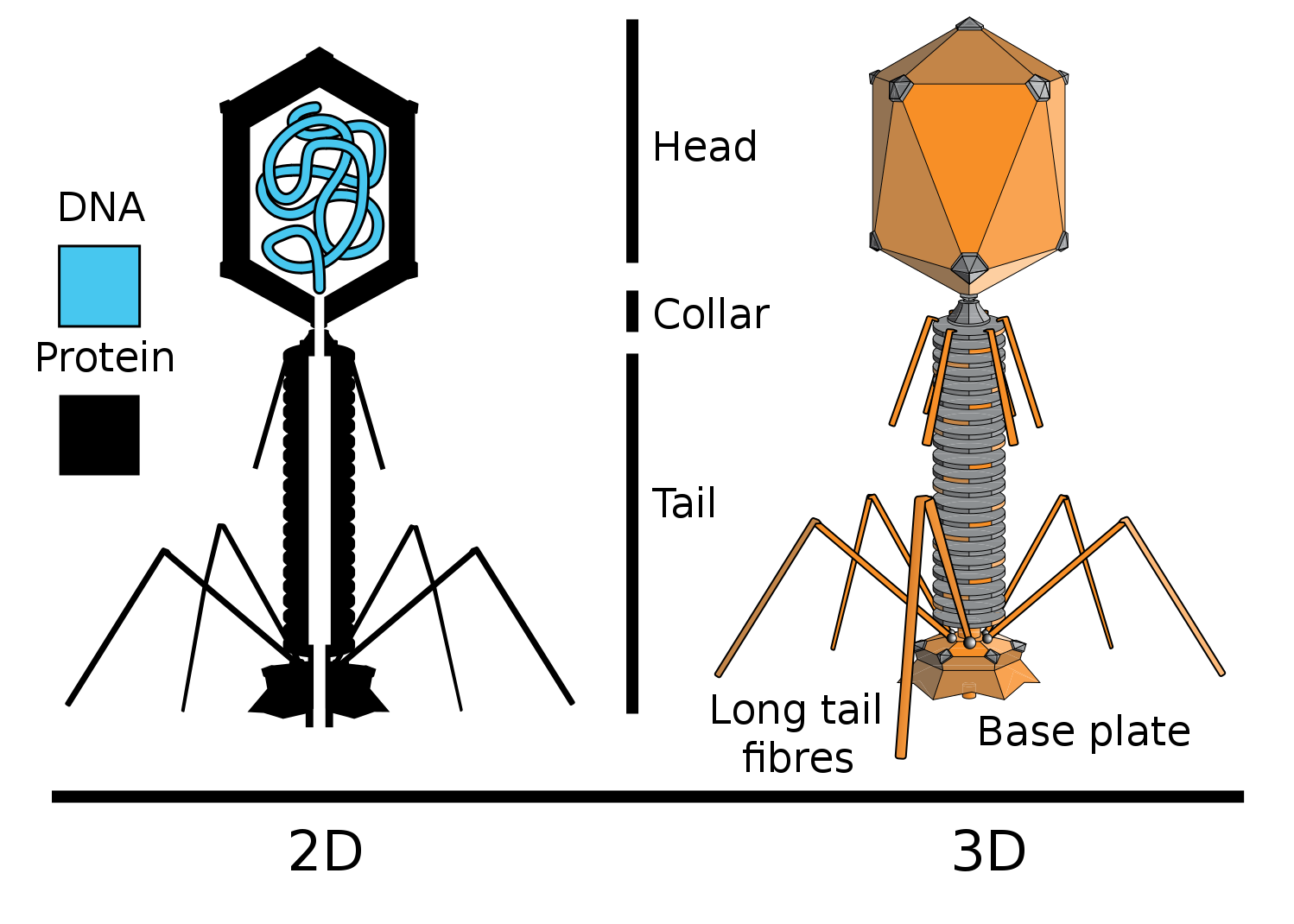
Actually what they did. They included radioactive phosphorus (P32) in the DNA of some bacteriophages, and the sulfur isotope in the proteins of others (S35). For this, some bacteria were grown on a medium with the addition of radioactive phosphorus in the composition of phosphate ion, others on a medium with the addition of radioactive sulfur in the composition of sulfate ion. Then bacteriophage T2 was added to these bacteria, which, multiplying in bacterial cells, included a radioactive label in its DNA (P is in DNA, but not in proteins), or proteins (S is in proteins, but not in DNA).
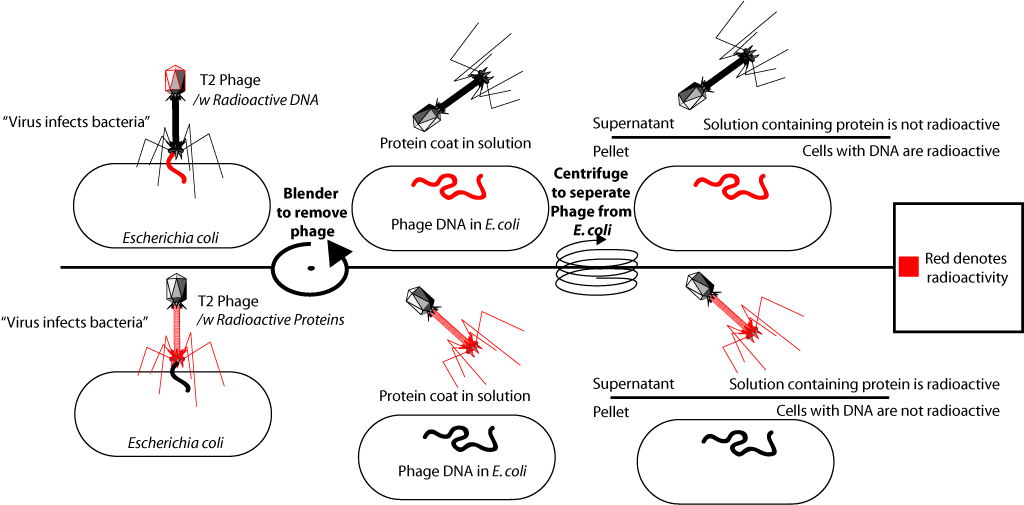
After isolation of the radioactively labeled bacteriophages, they were added to the culture of fresh (isotope-free) bacteria. What led to the infection of these bacteria. The bacteriophage joins the bacterial cell and "injects" its DNA. After that, the medium with bacteria was subjected to vigorous shaking in a special mixer (it was shown that in this case the phage membranes were separated from the surface of bacterial cells), and then infected bacteria were separated from the medium. When in the first experiment labeled with phosphorus-32 bacteriophages were added to the bacteria, it turned out that the radioactive label was in the bacterial cells. When, in the second experiment, bacteriophages labeled with sulfur-35 were added to the bacteria, the label was found in the fraction of the medium with protein shells, but it was not in the bacterial cells. This confirmed that the material that bacteria have been infected with is DNA. Since full viral particles containing viral proteins are formed inside infected bacteria, this experience was one of the decisive evidence that genetic information (information about the structure of proteins) is contained in DNA.
These discoveries greatly influenced many biologists of that time. Especially on the famous Chargaff. He believed that Avery had in fact discovered a 'new language', or at least showed where to look for it.
Chargaff began to look for a difference in the nucleotide composition and arrangement of nucleotides in DNA preparations obtained from various sources. And since there were no methods that could accurately give a chemical characterization of DNA, at that time ... he had to invent them. They were shown that the old tetranucleotide theory of the structure of nucleic acids is incorrect. DNA in different organisms in composition and structure are very different. At the same time, new facts were discovered that were not previously established for other natural polymers, namely, regularities in the ratio of individual bases in the composition of all the tested DNA. Now even schoolchildren know them as Chargaff rules.
- The amount of adenine is equal to the amount of thymine, and guanine is equal to cytosine: A = T, T = C.
- The number of purines is equal to the number of pyrimidines: A + G = T + C.
- It follows from the first and second. The number of bases with amino groups in position 6 is equal to the number of bases with keto groups in position 6: A + C = T + G.
We touched on the mechanism in the last article , so here I will not dwell on it.
Slowly we approached two legendary people who discovered the structure of DNA. Francis Creek and James Watson met for the first time in 1951. Watson then decided to take up the structure of DNA. As a biologist, he understood that when choosing a certain DNA structure, it is necessary to take into account the existence of some simple principle of doubling the DNA molecule embedded in its structure. Indeed, one of the most important properties of genes is the transmission of hereditary information.
A cry was used to create a theory of X-ray diffraction on spirals, which allows to determine whether the structure under study is in spiral conformation or not. DNA radiographs already existed at that time. They were received in London by Maurice Wilkins and Rosalind Franklin.
By the nature of the X-ray patterns of DNA, Watson and Crick realized that the structure under study was in a helical conformation. They also knew that the DNA molecule is a long linear polymer chain consisting of monomers and nucleotides. The phosphodeoxyribose backbone of this polymer is continuous, and nitrogenous bases are attached to the deoxyribose residues on the side. To build the models, it remained to solve the problem of how many chains of a linear polymer are packed in a compact structure.
Based on the X-ray diffraction pattern of the DNA, Watson and Crick suggested that the DNA molecule consists of two linear polynucleotide chains with a phosphodeoxyribose skeleton on the outside of the molecule and nitrogenous bases inside it. What later confirmed. It only remained to decide on the order of the location of the nitrogenous bases of the two chains inside the bipirale.
Considering possible combinations of nitrogenous base pairs, Watson found that adenine – thymine and guanine – cytosine pairs are the same size and are stabilized by hydrogen bonds. The rules of Chargaff were immediately explained: if a DNA adenine of one strand always binds to thymine of another strand, and a guanine always comes together with cytosine, then adenine in DNA should always be as much as thymine, and guanine - as many how much cytosine. It was also clear how the doubling of the DNA molecule should occur. Each strand is complementary to the other, and during the DNA replication process, the bispiral strand must disperse and a strand complementary to it must be completed on each polynucleotide strand. Here, too, there were several theories, but about them in a week, in the next article.
Coding Information
So, we know that DNA is a carrier of information, we know what it consists of. But how the information is encoded is still not clear.
Let's go from the task. DNA encodes 20 amino acids (we can say that 21, but so far we have not touched selenocysteine). Nucleotides have 4 options. That is, one nucleotide can encode 4 options, 2 - 16, 3 -64. It is logical to assume that the code is triplet (that is, the three bases encode one amino acid). You can read about experimental confirmation here . I am afraid that there is already a lot of history ...
Actually, we have 64 variants and 20 amino acids. Amino acids can be encoded by different codons. There are also start and stop codons from which reading begins.
Do not forget that the DNA is first read into RNA, which is already being read into a protein.
The table below is the correspondence of codons to RNA amino acids. Remember that there is no thymine in RNA, uracil instead.
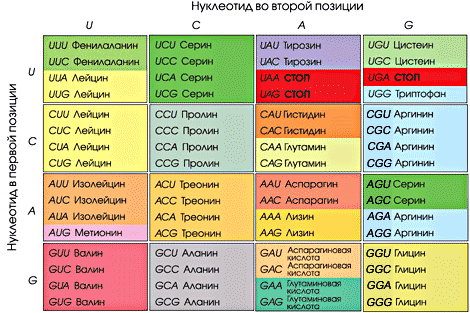
If you did not find the start codon in the table, look for the AUG. It encodes methionine and at the same time is the starting one. When broadcasting prokaryotic, plastid and mitochondrial genes, the starting amino acid is N-formylmethionine (this is just for reference)).
If we paint all the way from DNA to protein, we get something like that.
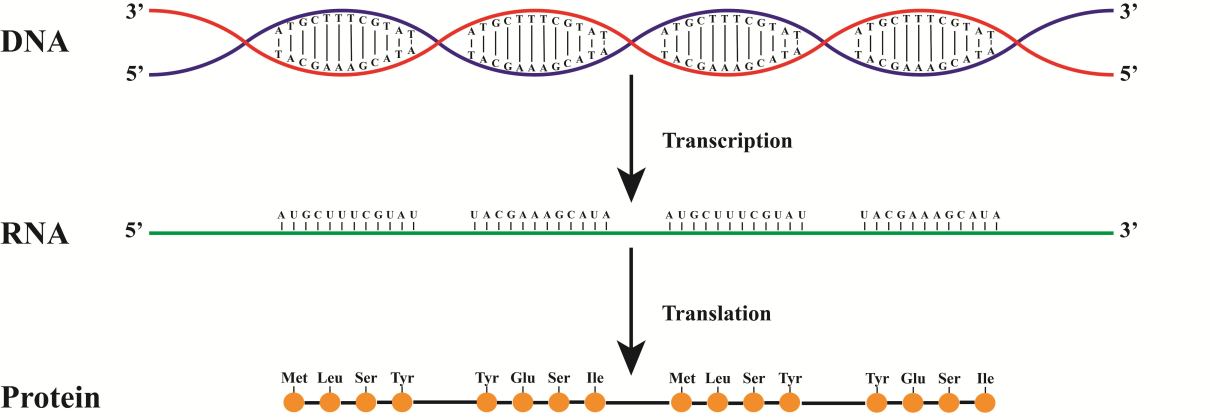
In this figure, the synthesis comes from the red chain. As a consequence, the RNA will coincide with the blue chain (do not forget about the replacement of T with Y)
As I said, each amino acid can be encoded by several codons. At first glance, this does not seem to be a particularly necessary side effect of redundancy in the number of codons. But he, in fact, quite an important role.
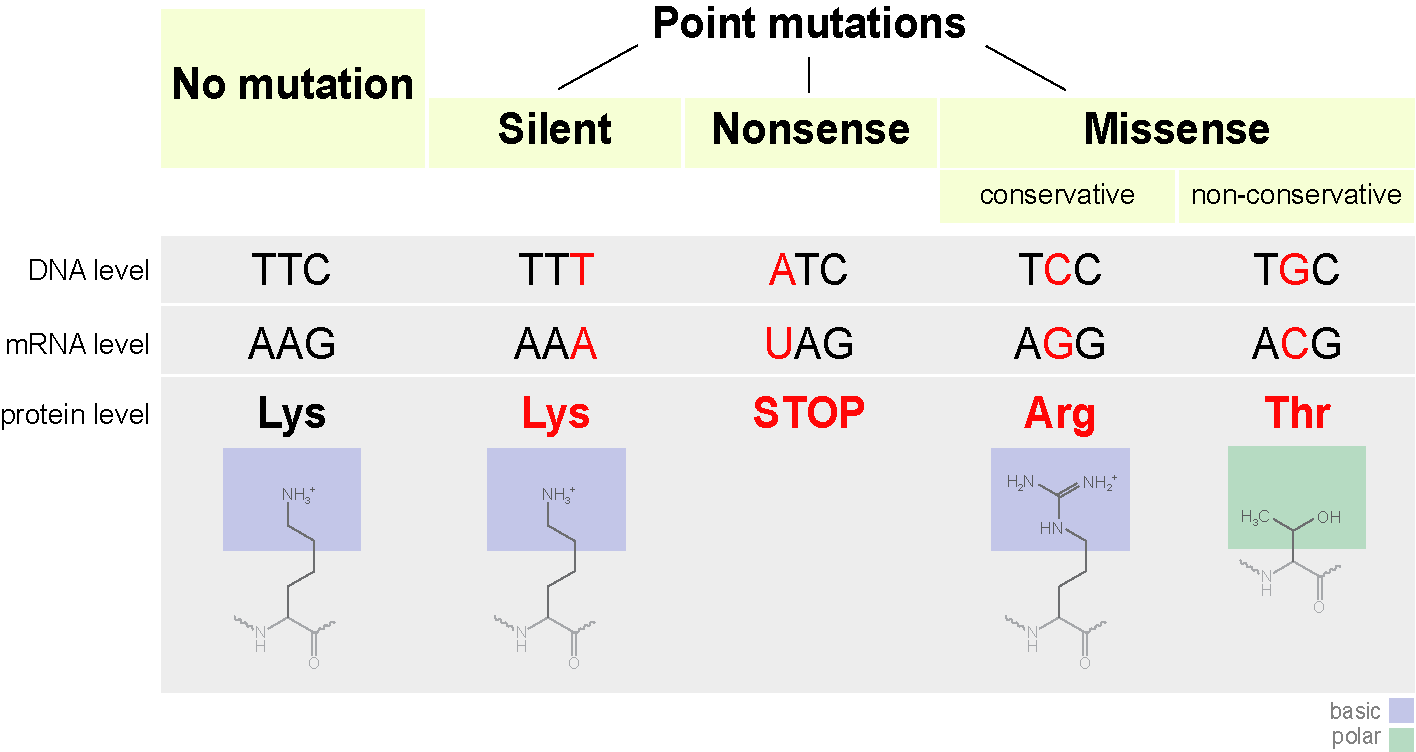
Here we will touch on a little mutation. They come in different types. From chromosomal, when whole pieces of chromosomes are removed from the genome, they change places, duplicate, to the point ones, when one nitrogenous base is replaced by another. Focus on point mutations.
What can point mutations lead to?
A codon can begin to encode another amino acid, which is not always scary. Such mutations are called missense mutations (that is, with a change of meaning). This can affect the structure of the protein. For example, if a positively charged amino acid is replaced by a negatively charged one, this can make the protein unstable, or lead to the fact that it coagulates into another conformation (yes, the linear sequence of amino acids usually collapses into a certain form) and cannot perform its functions (or better, it already smacks of evolution).
Specifically, hemoglobin S has a single nucleotide substitution (A for T) in the coding gene. As a result, the GAG triplet, coding for glutamate, is replaced with TG, which encodes valine. Hemoglobin S can also transport oxygen, but makes it worse than regular hemoglobin.
In the Hikari hemoglobin molecule, asparagine is substituted for lysine, but it is still good to carry oxygen.
As an example with loss of function, consider hemoglobin M. Another point mutation in the hemoglobin gene leads to a complete loss of function (histidine is changed to tyrosine in the active center).
By the way, folding protein looks like this, if you omit all the nuances.
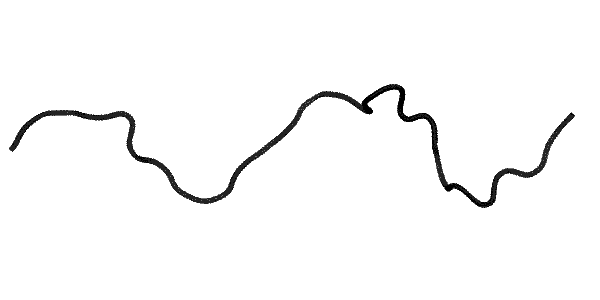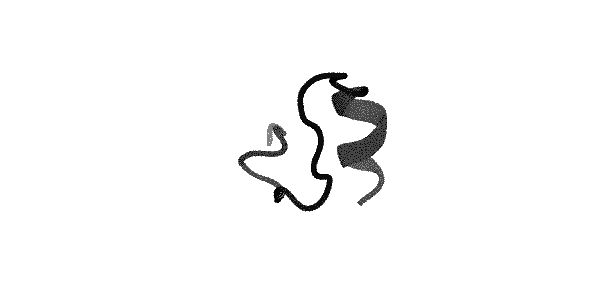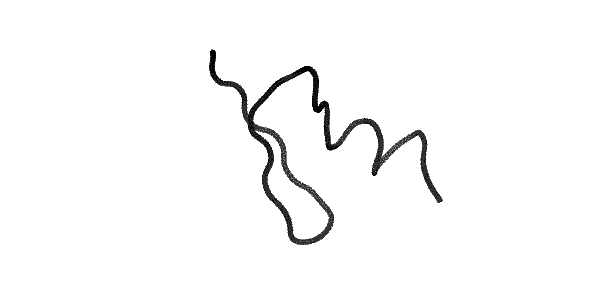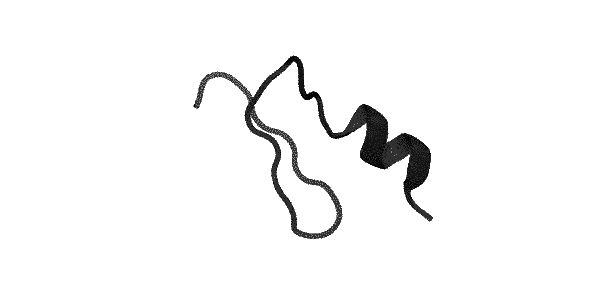
What else can happen?
Replacing a single nitrogenous base can also lead to the appearance of a stop codon in the center of the sequence, or vice versa, the stop codon at the end will disappear. The output will be either an incomplete circuit or an extremely long chain, which in any case will not be able to function normally. Such mutations are called nonsense.
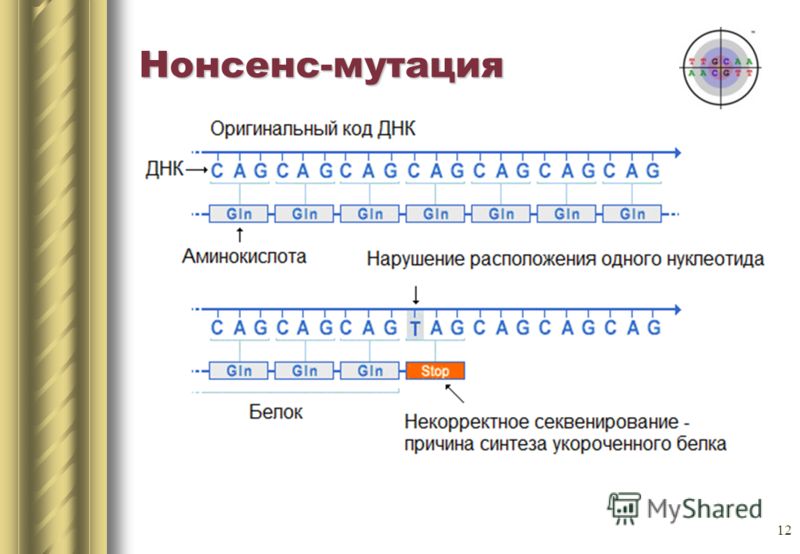
There is a third type of mutation - silence mutation. In fact, there is a change of codon to another, encoding the same amino acid. Protein properties do not change.
Let's summarize the general scheme.
In conclusion, I would like to tell you about one interesting feature. One amino acid can encode several codons. This we know. But what does it mean? The body immediately uses all codons for coding. But some more often, some less.
Let's compare a person and ... E. coli ( Escherichia coli ) in terms of frequency of use of cysteine encoding codons.
It is encoded by two codons UGU and UGC.
Human
UGU 10.6
UGC 12.6
E. coli (strain O127: H6)
UGU 19.1
UGC 0.0
The numbers are the occurrence of a triplet per thousand. It can be seen that we use both codons with approximately the same frequency, while E. coli almost does not use the UGC codon.
This feature needs to be remembered, especially when you are engaged in geno-engineering and you want to turn out the gene product of one organism in another. If a human gene, with frequent occurrence of a UGC codon, try to insert it into the E. coli of this strain - you will be disappointed. In the cell, amino acids are associated with transport RNAs, each of which corresponds to its own codon. So, the tRNA corresponding to the UGC codon will be extremely small, which will greatly slow down the synthesis.
If interested, here you can see the differences in the codon composition of different organisms.
The codon composition can be very different from that of organisms of different species, as well as of different strains. So in Escherichia coli O157: H7 EDL933 is more or less equally in terms of UGC and UGU. Or another example. The strains of the tubercle bacillus isolated in different countries also differ in code composition .
Summarize
This time there was a lot of history and relatively little biology. More of this will not happen. We talked about how it became clear that DNA is the carrier of information, as it is stored in the DNA itself. We talked about the redundancy of the gene code and what it leads to. Mutated by the mutations and the difference in the frequency of use of certain codons.
Next time let's talk about DNA replication.
PS: there will also be a story, but much less. I will try not to make such pauses in writing.
