Genome Browsers
Not the last role in bioinformatics is visualization. Scientists in this field work with huge volumes of information that it would be nice to somehow capture with a look and imagine in your head. A striking example of a visualization tool is the genome browser, which I want to talk about.
As many remember from the school biology course, the genome consists of a set of chromosomes, and the chromosome is two chains coiled. Each of the chains contains a nucleotide sequence with four types of nitrogenous bases - adenine (A), guanine (G), cytosine (C) and thymine (T). It is easy to determine the second from one chain, if you remember that adenine is paired with thymine (Antoshka-Timoshka), and guanine with cytosine (goose-chicken). Some sections of DNA are called genes, RNA is read from them, by which proteins are then encoded. Proteins are composed of 20 types of amino acids (plus a couple of exotic ones), each of which is encoded in three nucleotides.
A genome browser is a one-dimensional map that displays a nucleotide sequence (say, a chromosome or a single gene) with related information. Information is usually structured into blocks called tracks. For example, there may be a track with genes or with individual nucleotides. Individual entities on tracks are often called features.
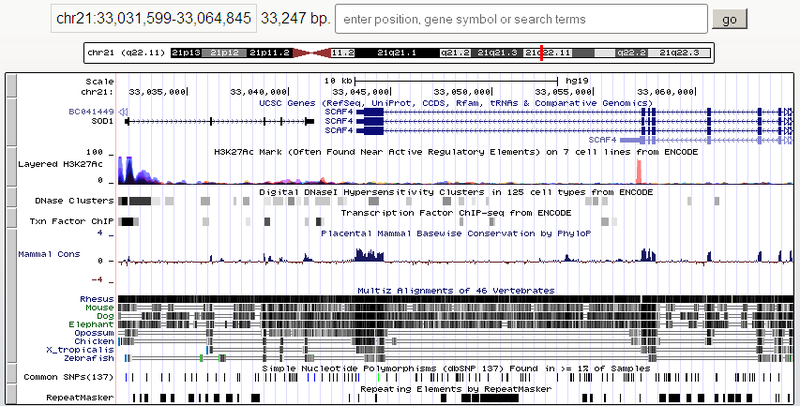
There are genome browsers tailored for small bacterial genomes, but a universal browser needs to show both the entire vertebrate chromosomes and individual nucleotides. The longest human chromosome ( first) contains about 250 million base pairs, that is, the scale should change about a million times. Of course, information is displayed differently on different scales. For example, in the picture above there is a track with the UCSC Genes genes, where the entire SOD1 gene and fragments of neighboring genes hit. At this scale, the exon-intron structure of the gene is displayed. The exons (those parts that remain in the RNA after splicing and in the future encode the protein) are indicated by shaded rectangles, and the introns (gaps between exons) are indicated by arrows that indicate the direction in which the gene is read (in this case, the SOD1 gene is located on a straight DNA strand, and BC041449 - on the back). And here is what a piece of the SOD1 gene looks like when zoomed in:

Here, the scale makes it possible to derive the amino acid sequence of those gene fragments that then encode the protein. Each amino acid corresponds to a specific letter of the Latin alphabet.
What else can you see on the genome browser? On the most detailed scale, you can see individual nucleotides, both on the direct and on the reverse helix of DNA: Each nucleotide has a standard color, so you can colorfully have fun even if the letters themselves no longer fit: If you roll back a little more, then the nucleotide composition can be judged on a special track GC content:



Here, the red color means that the nucleotides G and C in this place are less than 50%, and the blue color is more. You might think that A, C, G, T are just four equal states of a two-bit cell encoding genetic information, and the proportion of G and C does not mean anything interesting. However, GC base pairs form three hydrogen bonds, and AT only two. That is, GCs are stronger, they are more difficult to break and the enrichment of GC or AT bonds affects the chemical processes in a given region of DNA.
What else interesting can you see? Usually there are tracks with genomic variations that, for example, distinguish different people from each other. Variations are often expressed as point mutations, single-nucleotide substitutions ( SNPs)) Many of these mutations were found when comparing the results of sequencing genomes of different people and placed in special databases (for example, dbSNP):

There are not so few SNPs in the given fragment (19 for 356 nucleotides - more than 5%). However, many of them are synonymous. Since 20 variants of proteins are encoded from 4 3 = 64 variants of three nucleotides, some substitutions do not affect the resulting protein. Some substitutions fall into non-coding regions (for example, into introns), therefore they may also not affect the result (but they may also affect).
Another interesting thing is the comparison of the human genome with the genomes of other species. To do this, multiple alignment of genomes is made by non-trivial algorithms and also shown. The topmost picture of the post shows schematically the alignment of a person withrhesus macaque , mouse, dog, elephant, possum, chicken, frog (Xenopus tropicalis) and zebrafish zebrafish . The matching fragments are shown in dark. Note that the darkest regions are in the coding regions of genes. In the same picture, there is a graph of the conservatism of sites among mammals (Mammal cons), which also correlates. And here is the multiple alignment in an enlarged view:

Minus means that the nucleotide is in humans, but absent in another species. An orange vertical line (for example, in a line with a dog between two thymines) is the opposite. Above is the number of missing nucleotides (they themselves are not shown). The coding region is given in amino acid form, so synonymous substitutions are not visible. Chicken and fish apparently do not have a similar region at all. You can see how macaque looks like a person.
On the farthest scale, the karyotype of the chromosome becomes visible :

According to the karyotype, you can navigate if you remember, for example, in which band is your favorite gene that you are studying. Crossing in the middle is a centromere .
There are many other predefined tracks. Some browsers allow you to load tracks from the web using a special DAS protocol . Well, of course, genome browsers allow scientists to add their own (there are special file formats for this). Custom tracks can, for example, show areas of DNA binding to a specific protein (for example, with a transcription factor ), both predicted and obtained experimentally (for example, ChIP-Seq ). If, for example, you have sequenced your own genome, you can download the result and compare it with the reference and with known SNPs.
There are a lot of genome browsers. Only Wikipedia listedabout thirty, and that’s definitely not all. Many of them are specialized: tailored for a specific organism or a certain type of data. Of the popular desktop browsers, the Integrated Genome Browser and Integrative Genomic Viewer (as you can see, did not bother with the names). Both of them are Java applications, there is Java Web Start. Of course, it’s often more convenient to use the genome browser on the web. Most of the pictures above are made in the UCSC Genome Browser and Ensembl Genome Browser . Both of these browsers generate images on the server. There are more technically advanced solutions. Annoj

, for example, renders pictures on a client in canvas, receiving data in JSON from the server (a demonstration for the favorite weed of biologists - Arabidopsis). There is also JBrowse . It is unique in its own way, as it does not contain server code. Data on tracks and sequences are prepared in advance on the server in the form of static files, which the browser loads via AJAX. User files are processed through the File API .
An ideal genome browser does not exist. In my opinion, the main problem is the speed of work. This is especially noticeable on the web, although in the desktop there are delays. Some tracks at certain scales are either generated very slowly, or are turned off altogether. For visualization, it is necessary to grind a lot of information, which, perhaps, is not always presented in the optimal form. Therefore, if someone has a desire to do this, there is every chance to overcome competitors.

As many remember from the school biology course, the genome consists of a set of chromosomes, and the chromosome is two chains coiled. Each of the chains contains a nucleotide sequence with four types of nitrogenous bases - adenine (A), guanine (G), cytosine (C) and thymine (T). It is easy to determine the second from one chain, if you remember that adenine is paired with thymine (Antoshka-Timoshka), and guanine with cytosine (goose-chicken). Some sections of DNA are called genes, RNA is read from them, by which proteins are then encoded. Proteins are composed of 20 types of amino acids (plus a couple of exotic ones), each of which is encoded in three nucleotides.
A genome browser is a one-dimensional map that displays a nucleotide sequence (say, a chromosome or a single gene) with related information. Information is usually structured into blocks called tracks. For example, there may be a track with genes or with individual nucleotides. Individual entities on tracks are often called features.
There are genome browsers tailored for small bacterial genomes, but a universal browser needs to show both the entire vertebrate chromosomes and individual nucleotides. The longest human chromosome ( first) contains about 250 million base pairs, that is, the scale should change about a million times. Of course, information is displayed differently on different scales. For example, in the picture above there is a track with the UCSC Genes genes, where the entire SOD1 gene and fragments of neighboring genes hit. At this scale, the exon-intron structure of the gene is displayed. The exons (those parts that remain in the RNA after splicing and in the future encode the protein) are indicated by shaded rectangles, and the introns (gaps between exons) are indicated by arrows that indicate the direction in which the gene is read (in this case, the SOD1 gene is located on a straight DNA strand, and BC041449 - on the back). And here is what a piece of the SOD1 gene looks like when zoomed in:

Here, the scale makes it possible to derive the amino acid sequence of those gene fragments that then encode the protein. Each amino acid corresponds to a specific letter of the Latin alphabet.
What else can you see on the genome browser? On the most detailed scale, you can see individual nucleotides, both on the direct and on the reverse helix of DNA: Each nucleotide has a standard color, so you can colorfully have fun even if the letters themselves no longer fit: If you roll back a little more, then the nucleotide composition can be judged on a special track GC content:



Here, the red color means that the nucleotides G and C in this place are less than 50%, and the blue color is more. You might think that A, C, G, T are just four equal states of a two-bit cell encoding genetic information, and the proportion of G and C does not mean anything interesting. However, GC base pairs form three hydrogen bonds, and AT only two. That is, GCs are stronger, they are more difficult to break and the enrichment of GC or AT bonds affects the chemical processes in a given region of DNA.
What else interesting can you see? Usually there are tracks with genomic variations that, for example, distinguish different people from each other. Variations are often expressed as point mutations, single-nucleotide substitutions ( SNPs)) Many of these mutations were found when comparing the results of sequencing genomes of different people and placed in special databases (for example, dbSNP):

There are not so few SNPs in the given fragment (19 for 356 nucleotides - more than 5%). However, many of them are synonymous. Since 20 variants of proteins are encoded from 4 3 = 64 variants of three nucleotides, some substitutions do not affect the resulting protein. Some substitutions fall into non-coding regions (for example, into introns), therefore they may also not affect the result (but they may also affect).
Another interesting thing is the comparison of the human genome with the genomes of other species. To do this, multiple alignment of genomes is made by non-trivial algorithms and also shown. The topmost picture of the post shows schematically the alignment of a person withrhesus macaque , mouse, dog, elephant, possum, chicken, frog (Xenopus tropicalis) and zebrafish zebrafish . The matching fragments are shown in dark. Note that the darkest regions are in the coding regions of genes. In the same picture, there is a graph of the conservatism of sites among mammals (Mammal cons), which also correlates. And here is the multiple alignment in an enlarged view:

Minus means that the nucleotide is in humans, but absent in another species. An orange vertical line (for example, in a line with a dog between two thymines) is the opposite. Above is the number of missing nucleotides (they themselves are not shown). The coding region is given in amino acid form, so synonymous substitutions are not visible. Chicken and fish apparently do not have a similar region at all. You can see how macaque looks like a person.
On the farthest scale, the karyotype of the chromosome becomes visible :

According to the karyotype, you can navigate if you remember, for example, in which band is your favorite gene that you are studying. Crossing in the middle is a centromere .
There are many other predefined tracks. Some browsers allow you to load tracks from the web using a special DAS protocol . Well, of course, genome browsers allow scientists to add their own (there are special file formats for this). Custom tracks can, for example, show areas of DNA binding to a specific protein (for example, with a transcription factor ), both predicted and obtained experimentally (for example, ChIP-Seq ). If, for example, you have sequenced your own genome, you can download the result and compare it with the reference and with known SNPs.
There are a lot of genome browsers. Only Wikipedia listedabout thirty, and that’s definitely not all. Many of them are specialized: tailored for a specific organism or a certain type of data. Of the popular desktop browsers, the Integrated Genome Browser and Integrative Genomic Viewer (as you can see, did not bother with the names). Both of them are Java applications, there is Java Web Start. Of course, it’s often more convenient to use the genome browser on the web. Most of the pictures above are made in the UCSC Genome Browser and Ensembl Genome Browser . Both of these browsers generate images on the server. There are more technically advanced solutions. Annoj

, for example, renders pictures on a client in canvas, receiving data in JSON from the server (a demonstration for the favorite weed of biologists - Arabidopsis). There is also JBrowse . It is unique in its own way, as it does not contain server code. Data on tracks and sequences are prepared in advance on the server in the form of static files, which the browser loads via AJAX. User files are processed through the File API .
An ideal genome browser does not exist. In my opinion, the main problem is the speed of work. This is especially noticeable on the web, although in the desktop there are delays. Some tracks at certain scales are either generated very slowly, or are turned off altogether. For visualization, it is necessary to grind a lot of information, which, perhaps, is not always presented in the optimal form. Therefore, if someone has a desire to do this, there is every chance to overcome competitors.
