DNA through the eyes of a programmer
From the translator: Since I am not a biologist, inaccuracies in the translation of terms are possible (and not only :). The original is here .
These are just programmer’s thoughts about DNA. I am not a molecular geneticist.
Is here . It is not joke. The source can be viewed using a great Perl script set called " Ensembl ". The human genome occupies approximately 3 gigabytes, which can be reduced to 750 megabytes, if you discard the husks. A bit sad that this is just 2.8 Mozilla Firefox browsers .
DNA is more likely not like C source code, but rather a byte-code for a virtual machine called the “cell nucleus”. It is extremely doubtful that there are source codes that can be compiled into this bytecode: what we see is all that we have.
 DNA language is digital, but not binary. Binary code uses 0 and 1 (therefore it is called binary), DNA uses 4 values: T, C, G and A.
DNA language is digital, but not binary. Binary code uses 0 and 1 (therefore it is called binary), DNA uses 4 values: T, C, G and A.
While the binary byte consists mainly of 8 binary digits, the DNA is “byte” (called the codon ) contains 3 characters. And since each character can have one of four values, the DNA codon has 64 possible values, in contrast to the 256 values of the binary byte.
A typical example of a DNA codon is “GCC”, which encodes the amino acid Alanine. The combination of a large number of these amino acids is called a polypeptide or protein, and is a chemically active component of all living things. Read more about codons.
The code of dynamic link libraries (.so on Unix systems, .dll on Windows) cannot use static addresses internally, since this code can be located in different parts of memory in different situations. DNA also has a similar function called "code transposition":
Most cells use only a very small fraction of the estimated 20,000-30000 genes of the human genome — which is reasonable, since the liver cell does not need the DNA code that makes up the neurons.
But in view of the fact that all cells contain a complete copy (“distributive”) of the genome, a system is needed that will allow to mark
That is why the " stem cells " are now so popular - this type of cell has the ability to turn into anything. The code is not yet
More precisely, in stem cells, everything is not included - they are not both liver cells and neurons at the same time. Cells can be represented as a finite state machine, starting from the state of a stem cell. During the life cycle of a cell, during which it can divide (
Each cell can make (or be prompted to make) a decision about its future; each of these decisions makes it more specialized. These solutions are saved when dividing using transcription factors and modifying the spatial storage of DNA (steric effects).
The liver cell in general cannot function as a skin cell, despite the fact that it contains all the necessary genetic instructions for this. However, there are indications that cells can be “bred” up the hierarchy, making them pluripotent.
Despite the fact that full-fledged changes in the DNA in the body rarely occur within a single generation, significant amendments are made by activating or deactivating parts of our genome without changing the code itself.
To make an analogy, this is similar to the Linux kernel, which at boot time detects the processor on which it is running, and disables parts of its binary code in the case (for example) if it is running on a uniprocessor system. This is not just something like
Similarly, as the embryo develops in the womb, its DNA is substantially edited to reduce its growth rate and placenta size. In this way, the opposing interests of the father (“big strong child”) and the mother (“survive the pregnancy”) are balanced. Such “imprinting” can occur only inside the mother, since the father’s genome has no idea about the size of the mother.
Recently it has also become clear that the metabolic status of parents affects the likelihood of a long life, the onset of cancer and diabetes in their grandchildren , which is quite logical, because survival in a poor food climate may require a completely different metabolic strategy in comparison with life in the food environment is enough.
The mechanisms behind epigenetics and imprinting, “methylation”, attach methyl groups to DNA to switch the activation status of individual sections, as well as modification of histones, which can turn off DNA segments, which leads to deactivation of these sections.
Some of these DNA edits are inherited and transmitted to the offspring; other forms affect only a single animal.
This area of science is still developing rapidly, and it may turn out that our DNA is much more dynamic than we thought.
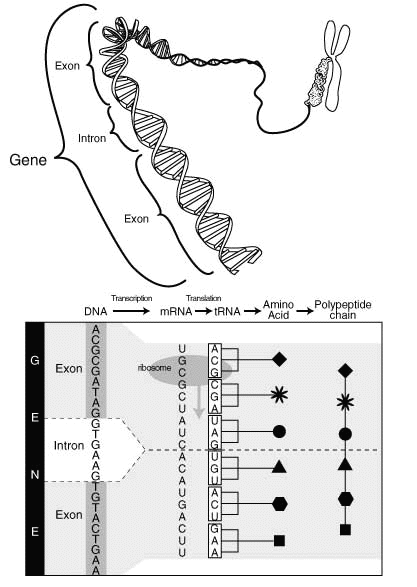 The genome is clogged with old copies of genes and failed experiments in the recent past (about half a million years ago). This code is still in DNA, but inactive. It is called "pseudogenic."
The genome is clogged with old copies of genes and failed experiments in the recent past (about half a million years ago). This code is still in DNA, but inactive. It is called "pseudogenic."
Moreover, 97% of your DNA is commented out. DNA is linear and readable from start to finish. Those sections that do not need to be decoded are clearly marked, just like the comments in C. The remaining 3%, which are used for their intended purpose, form the so-called “exons”. Comments that are “between” are called “introns”.
These comments are charming in their own way. Like C comments, they have start markers (like
However, due to subsequent cutting, some glue is required to which the code will be connected before the comment with the code after the comment, which makes these comments look more like comments in HTML that are somewhat longer: - end.
So, in fact, a gap of DNA with exons and introns may look something like this:
With the beginning of the comment, everything is simple, followed by a huge amount of non-coding DNA. Somewhere near the end of the comment is a branch site, which means the comment will end soon. After it comes a little more comment, and then a real comment limiter.
Directly cutting out the comments occurs after the DNA is transcribed into RNA and is performed by twisting the comments into a loop and thus combining pieces of the present code. After that, RNA is cut at the branch sites near the end of the comment, then the “donor” (the beginning of the comment) and the “acceptor” (the end) are linked to each other.
So what are these comments for? This topic is for a holivar of power comparable to vim / emacs. When comparing different species, we learn that some introns contain fewer differences in code than neighboring exons. This suggests that comments perform some important function.
There are many plausible explanations for the existence of a huge amount of non-coding DNA - one of which, the most attractive (for a programmer), involves folding propensity. DNA should be stored in a tightly folded form, but not all codes in DNA can handle this fully.
In part, this may resemble RLL or MFM coding. On a hard disk, bits are encoded by the presence or absence of polarity inversion. With the naive coding method, we would write 0 as “no inversion” and 1 as “inversion”.
Encode the 000000 sequence with this method is extremely easy - it is enough to leave the magnetic phase unchanged for several micrometers. However, when decoding, we will encounter ambiguity - how many micrometers have we read? Is this equal to 6 zeros or 5? To prevent this problem, the data is recorded in such a way that such continuous sections without inversion will not occur.
If we see the “no inversion, no inversion, inversion, inversion” sequence on the disk, we can be sure that it corresponds to “0011” - it is extremely unlikely that our reading process is so inaccurate that we can interpret it as “00011” or "00111". Thus, we need to insert delimiters to prevent an insufficient number of transitions. This approach is called “Run Length Limiting (RLL)” on magnetic media.
We emphasize once again that sometimes it is necessary to insert transitions (inversions) in order for the data storage to be reliable. It is possible that the introns perform a similar function, allowing the end code to fold appropriately.
Anyway, molecular biology is a minefield! Controversy and criticism are raging around options with exciting names like “early introns” or “late introns”, as well as such important words as “stem-loop potential”. I think it is better to let this debate rage on.
Update from the author: ten years later, this debate has not yet subsided. Now it is clear that “junk DNA” is an incorrect name, but as far as its immediate function is concerned, consensus has not been reached.
As in the case of unix, cells are not “spawned” - they divide. All cells in your body have a beginning in the form of an egg, which has since been divided countless times. Like the processes, both halves that arise as a result are
As in unix, significant problems arise when cells continue to
A cell cannot clone itself until extremely stringent conditions are met - the default security settings . And only if all these defense mechanisms fail, a tumor may develop. As in computer security, it is difficult to find a balance between security (“no cells can share”) and usability.
You can compare this with a known problem with a stop , first described by computer science founder Alan Turing. It seems that to predict whether a program will ever end is just as easy as creating a functional genome that will never lead to cancer?
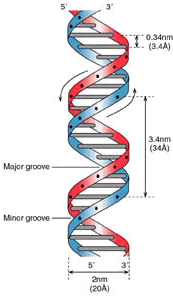 Each DNA helix is redundant in itself - it looks like a twisted ladder in which each step contains two bases - hence the word “pair of bases”. If one of these bases is missing, it can be removed from the base on the other side of the "step". T is always associated with A, C is always with G. So we can argue that the genome is mirrored inside the helix, like RAID-1.
Each DNA helix is redundant in itself - it looks like a twisted ladder in which each step contains two bases - hence the word “pair of bases”. If one of these bases is missing, it can be removed from the base on the other side of the "step". T is always associated with A, C is always with G. So we can argue that the genome is mirrored inside the helix, like RAID-1.
Moreover, the chromosomes are always presented in duplicate - one from each parent, with one notable exception - the Y chromosome, which occurs only in males. Although the details are somewhat more complicated, we can assume that we have two copies of most genes. In the event that one is damaged or mutated into a useless state, we still have a second, independent copy. We usually call this failover.
During the interaction within the cell, proteins rely on the characteristics of each other. In 2001-2002, studies were published that proteins that interact with a large number of other proteins are not able to evolve, or at least do it very slowly (Nature Journal, June 28, 2001, and M. Kimura, T. Ohta in Science journal, April 26, 2002).
The authors of the research suggest that this is due to the huge number of internal dependencies that prevent a change in the protein “contract”. It was also noted that evolution is still happening, but very slowly, since both sides of the dependency must simultaneously evolve in a compatible way.
Recently, during the conversation, someone suggested that it would be really cool to break the genome and compromise by inserting a code that would copy itself to other genomes, using the host body as a transport. "Just like the Nimda worm!" He
soon realized that this is what biological viruses have been doing for millions of years. And they are very good at it.
A large number of such viruses have become an integral part of our genome and ride with all of us. To do this, they have to hide from the virus scanner, which seeks to detect someone else's, malicious code and prevent its penetration into the DNA.
Central dogma:
At the dawn of the discoveries of the foundations of genetics, scientists had to deal with a variety of chemicals, the interrelationship of which was not obvious. The moment when it became clear from what it turns out, was proclaimed a great triumph and received the name "Central Dogma."
This dogma tells us that DNA is needed to get RNA, and RNA is needed to get proteins, just as it turns from an .c file into an .o object file, which can then be compiled into an executable file (a.out / exe) . In addition, she tells us that this is the only order in which the information flow follows.
And yet, the Central Dogma has recently faded a little. As with any billion-year project development, there were a lot of hacks, because of which information sometimes flows in the opposite direction. Sometimes RNA patches DNA, and it happens that DNA is modified by proteins called previously created.
But, in general, the dependencies are clear, so the Central Dogma still remains important.
Making fraud with DNA is pretty simple. There are companies to which you can send a file with DNA written in ASCII characters, and they synthesize relevant materials for you. We can even make up the DNA of the embryos of animals or plants.
It is much more difficult to patch an “executable file” right at runtime, and any programmer will confirm this. The same is with the genome. To change a running copy (of a person, for example), you need to edit every instance of the gene that you want to patch in the body.
For many years, medical science has been trying to patch people suffering from severe combined immunodeficiency (SCID, Severe Combined Immunodefeciency) - an extremely unpleasant disease that essentially disables the immune system, making the patient seriously ill. For a long time it became clear exactly what letters in the DNA must be corrected for the healing of these people.
Many attempts have been made to patch the “working” people with the help of viruses that would insert a new DNA into living organisms, but this approach has proved too complicated. The genome is extremely well protected to make such a simple approach — the cells store their code more reliably than Microsoft!
Nevertheless, a suitable virus was recently found that is able to break through the protection and correct DNA breaks, which, like,heals the sick .
As bugs are fixed in computer programs, we often introduce new bugs. The genome is replete with similar bugs. African Americans are immune to malaria, but instead are susceptible to sickle-cell anemia:
And this is not the only example of such a regression.
Like a computer drive, DNA (and its intermediate form, RNA) can be damaged. To prevent common “one-bit errors”, protein coding in DNA symbols is redundant. There are 4 symbols of RNA: U, C, G, and A — in other words, a “byte” is 2 bits long. Three characters correspond to the amino acid.
Theoretically, 6 bits could correspond to 64 amino acids, however there are only 20 standard amino acids. For example, UCU, UCC, UCA and UCG - encode the same serine, whereas only UGG corresponds to tryptophan.
Then, it turns out that some of the most likely "typos" (UCU -> UCC) in the code will lead to the synthesis of the same amino acid. Read more about this phenomenon in the book " Metamagical Themas"by Douglas Hofstadter (Douglas Hofstadter).
Some parts of the code are sacred. We may not remember who or why wrote it, we just know that it works. The guy who wrote it may have long left the company. With this code it is better not to get involved.
The concept of molecular hours is known in DNA. Some parts of the genome are actively changing, and some are inviolable. An excellent example of the latter is the histone genes H3 and H4.
These genes underlie how the genome is stored, and therefore are of paramount importance. Any failure in their code leads to a non-viable organism.
Therefore, we can assume that this code does not change in haste, and, apparently, the way it is. H3 and H4 genes have zeroeffective mutation rate in humans. But it goes even further. This code is common to everyone, starting with a person, ending with grass or mold.
Apparently, there are two ways by which the genome can verify that there is no code mutation. The first method was described earlier: to use amino acids, the coding of which is very redundant, which will allow to get the same result even in the case of "typos".
The second way is that the genes can be copied at the beginning or end of the cell's reproduction process, which provides more or less favorable conditions for copying. In addition, a huge number of similar conditions are used.
It seems that H3 and H4 were very carefully worked out, since they allow a lot of "synonymous" changes, which, thanks to the clever techniques described above, do not lead to a change in the result.
This sequence obviously describes the 8-bit values 1, 2, and 3. I added spaces to show where the byte begins and ends. Many serial devices use stop and start bits to indicate where you need to start reading. If we move this sequence a little:
it will suddenly begin to denote 2, 4, 6! To avoid this, signals are used in the DNA that are generated that tell the cell where to start reading. It is curious that there are parts of the genome that you can start reading from several starting points, and each time get a useful (but different) result. That's what I call a cool hack!
Each way of reading such a piece of DNA is called an open reading frame and there are usually 6 of them, 3 in each direction.
And yet, DNA is not like a programming language. She really is not. However, there are some big similarities. You can consider each cell as a central processor, each of which runs its own OS kernel. Each cell has a complete copy of the core, but only activates the corresponding parts - its own modules and drivers, so to speak.
When a cell needs to do something (“call a function”), it finds the necessary part of the genome and transcribes it into RNA. The RNA is then translated into a sequence of amino acids, which together give the protein that was encoded in the DNA. And now the coolest part!
This protein will be labeled with a shipping address. It is presented in the form of a marker consisting of several amino acids, which tells the cell exactly where to deliver this protein. Inside the cell there is a whole mechanism that carries out these instructions and is able to deliver the protein to an address that may even be outside the cell itself.
After delivery, this instruction comes off and several post-processing steps are carried out, which can, for example, activate protein, because transporting active protein through places for which it is not intended is not the best idea.
Organisms usually begin their existence from a single cell, which, as mentioned earlier, contains two full-fledged copies of the genome. A peculiar tar file in which all files are already unpacked, ready to use. What next?
Meet the homeobox. Cells must be copied and each must be assigned its purpose of existence. Genes that contain a homeobox begin to build a “top-down” dependency chain, which means “start from the head.” In order for this to work, a chemical gradient is formed, thanks to which the cells can sense where they are and determine their further actions required for the formation of the head or for the formation of the notochord.
First discovered in 1983, homeobox genes are currently one of the most exciting areas to explore. Note the curious fact that, like the makefile, the “HOX” genes are only responsible for the inclusion of different things in other genes and in themselves form nothing.
It seems that the “syntax” of the homeobox is “sacred” in the sense described above. What happens if you copy the “HOX” part of the mouse gene, which is the “leg selector” to the Drosophila homeoobox?
The genomes of fruit flies and humans branched not millions but hundreds of millions of years ago. And yet you can copy the plots (“selectors” in the language of genetics) makefiles, and they will work. Please note that the “growing legs” procedure in a fruit fly is completely different from a mouse, but the “selector” is able to correctly run the necessary instructions.
All living organisms have DNA, sometimes organized into several chromosomes (“libraries”), sometimes only one (usually looped out in such cases). This also applies to most bacteria. And in the neighborhood of this large main genome, such bacteria often contain “plasmids” - tiny DNA rings with specific functions.
These plasmids are partly compatible with different species and are transmitted horizontally using a variety of mechanisms. In this way, for example, even non-identical bacteria can “learn” antibiotic resistance from each other.
When compared to the world of programming, plasmids are not a voluntary acquisition, and are similar to loading .so using LD_PRELOAD or its equivalent on other platforms. Indeed, plasmids are often injected for research purposes. They can be introduced into any kind of bacteria and immediately get to work.
Plasmids copy themselves independently of the main chromosome, therefore they become a constant component of the bacterium. To achieve this, a plasmid possesses a gene with the great name “replication origin”, which is triggered when a cell is about to divide.
If you are a hammer, you will see a nail in everything.
These are just programmer’s thoughts about DNA. I am not a molecular geneticist.
Source
Is here . It is not joke. The source can be viewed using a great Perl script set called " Ensembl ". The human genome occupies approximately 3 gigabytes, which can be reduced to 750 megabytes, if you discard the husks. A bit sad that this is just 2.8 Mozilla Firefox browsers .
DNA is more likely not like C source code, but rather a byte-code for a virtual machine called the “cell nucleus”. It is extremely doubtful that there are source codes that can be compiled into this bytecode: what we see is all that we have.
 DNA language is digital, but not binary. Binary code uses 0 and 1 (therefore it is called binary), DNA uses 4 values: T, C, G and A.
DNA language is digital, but not binary. Binary code uses 0 and 1 (therefore it is called binary), DNA uses 4 values: T, C, G and A. While the binary byte consists mainly of 8 binary digits, the DNA is “byte” (called the codon ) contains 3 characters. And since each character can have one of four values, the DNA codon has 64 possible values, in contrast to the 256 values of the binary byte.
A typical example of a DNA codon is “GCC”, which encodes the amino acid Alanine. The combination of a large number of these amino acids is called a polypeptide or protein, and is a chemically active component of all living things. Read more about codons.
Position Independent Code
The code of dynamic link libraries (.so on Unix systems, .dll on Windows) cannot use static addresses internally, since this code can be located in different parts of memory in different situations. DNA also has a similar function called "code transposition":
Almost half of the human genome consists of transposable (or "mobile") genetic elements. These elements were first discovered in the 1940s by Dr. Barbara McClintock when studying the specific patterns of inheritance found in the colors of Indian corn. The idea of mobile DNA is that some areas are unstable and “transposable”, that is, they can move - inside and between chromosomes.
Conditional compilation
Most cells use only a very small fraction of the estimated 20,000-30000 genes of the human genome — which is reasonable, since the liver cell does not need the DNA code that makes up the neurons.
But in view of the fact that all cells contain a complete copy (“distributive”) of the genome, a system is needed that will allow to mark
#ifdefunnecessary things. And that's how it works. The genetic code is teeming with directives #if/#endif. That is why the " stem cells " are now so popular - this type of cell has the ability to turn into anything. The code is not yet
#ifdefen, so to speak.More precisely, in stem cells, everything is not included - they are not both liver cells and neurons at the same time. Cells can be represented as a finite state machine, starting from the state of a stem cell. During the life cycle of a cell, during which it can divide (
fork()) many times, it specializes. Each specialization can be considered as a choice of a branch in a tree. Each cell can make (or be prompted to make) a decision about its future; each of these decisions makes it more specialized. These solutions are saved when dividing using transcription factors and modifying the spatial storage of DNA (steric effects).
The liver cell in general cannot function as a skin cell, despite the fact that it contains all the necessary genetic instructions for this. However, there are indications that cells can be “bred” up the hierarchy, making them pluripotent.
Epigenetics and imprinting: patches during execution
Despite the fact that full-fledged changes in the DNA in the body rarely occur within a single generation, significant amendments are made by activating or deactivating parts of our genome without changing the code itself.
To make an analogy, this is similar to the Linux kernel, which at boot time detects the processor on which it is running, and disables parts of its binary code in the case (for example) if it is running on a uniprocessor system. This is not just something like
if (numcpus > 1), this code is really replaced by nop-ami. It is imperative that nop-sh is happening in memory, and not in an image on disk.Similarly, as the embryo develops in the womb, its DNA is substantially edited to reduce its growth rate and placenta size. In this way, the opposing interests of the father (“big strong child”) and the mother (“survive the pregnancy”) are balanced. Such “imprinting” can occur only inside the mother, since the father’s genome has no idea about the size of the mother.
Recently it has also become clear that the metabolic status of parents affects the likelihood of a long life, the onset of cancer and diabetes in their grandchildren , which is quite logical, because survival in a poor food climate may require a completely different metabolic strategy in comparison with life in the food environment is enough.
The mechanisms behind epigenetics and imprinting, “methylation”, attach methyl groups to DNA to switch the activation status of individual sections, as well as modification of histones, which can turn off DNA segments, which leads to deactivation of these sections.
Some of these DNA edits are inherited and transmitted to the offspring; other forms affect only a single animal.
This area of science is still developing rapidly, and it may turn out that our DNA is much more dynamic than we thought.
Dead code, bloated code, comments (garbage in DNA)
 The genome is clogged with old copies of genes and failed experiments in the recent past (about half a million years ago). This code is still in DNA, but inactive. It is called "pseudogenic."
The genome is clogged with old copies of genes and failed experiments in the recent past (about half a million years ago). This code is still in DNA, but inactive. It is called "pseudogenic." Moreover, 97% of your DNA is commented out. DNA is linear and readable from start to finish. Those sections that do not need to be decoded are clearly marked, just like the comments in C. The remaining 3%, which are used for their intended purpose, form the so-called “exons”. Comments that are “between” are called “introns”.
These comments are charming in their own way. Like C comments, they have start markers (like
/*) and end markers (*/). But they have a more complex structure. After all, as you remember, DNA is like a ribbon, respectively, comments need to be cut out physically! The beginning of a comment is almost always indicated by the letters “GT”, which can be compared with /*, the ending is indicated by the symbols “AG”, which, respectively, are similar */. However, due to subsequent cutting, some glue is required to which the code will be connected before the comment with the code after the comment, which makes these comments look more like comments in HTML that are somewhat longer: - end.
So, in fact, a gap of DNA with exons and introns may look something like this:
real code <! - bla bla bla bla bla ---- bla -> real code
| | | | | |
exon 1 donor * intron 1 branch acceptor ** exon 2
* start of comment
** end of comment
With the beginning of the comment, everything is simple, followed by a huge amount of non-coding DNA. Somewhere near the end of the comment is a branch site, which means the comment will end soon. After it comes a little more comment, and then a real comment limiter.
Directly cutting out the comments occurs after the DNA is transcribed into RNA and is performed by twisting the comments into a loop and thus combining pieces of the present code. After that, RNA is cut at the branch sites near the end of the comment, then the “donor” (the beginning of the comment) and the “acceptor” (the end) are linked to each other.
So what are these comments for? This topic is for a holivar of power comparable to vim / emacs. When comparing different species, we learn that some introns contain fewer differences in code than neighboring exons. This suggests that comments perform some important function.
There are many plausible explanations for the existence of a huge amount of non-coding DNA - one of which, the most attractive (for a programmer), involves folding propensity. DNA should be stored in a tightly folded form, but not all codes in DNA can handle this fully.
In part, this may resemble RLL or MFM coding. On a hard disk, bits are encoded by the presence or absence of polarity inversion. With the naive coding method, we would write 0 as “no inversion” and 1 as “inversion”.
Encode the 000000 sequence with this method is extremely easy - it is enough to leave the magnetic phase unchanged for several micrometers. However, when decoding, we will encounter ambiguity - how many micrometers have we read? Is this equal to 6 zeros or 5? To prevent this problem, the data is recorded in such a way that such continuous sections without inversion will not occur.
If we see the “no inversion, no inversion, inversion, inversion” sequence on the disk, we can be sure that it corresponds to “0011” - it is extremely unlikely that our reading process is so inaccurate that we can interpret it as “00011” or "00111". Thus, we need to insert delimiters to prevent an insufficient number of transitions. This approach is called “Run Length Limiting (RLL)” on magnetic media.
We emphasize once again that sometimes it is necessary to insert transitions (inversions) in order for the data storage to be reliable. It is possible that the introns perform a similar function, allowing the end code to fold appropriately.
Anyway, molecular biology is a minefield! Controversy and criticism are raging around options with exciting names like “early introns” or “late introns”, as well as such important words as “stem-loop potential”. I think it is better to let this debate rage on.
Update from the author: ten years later, this debate has not yet subsided. Now it is clear that “junk DNA” is an incorrect name, but as far as its immediate function is concerned, consensus has not been reached.
fork() and fork bombs (tumors)
As in the case of unix, cells are not “spawned” - they divide. All cells in your body have a beginning in the form of an egg, which has since been divided countless times. Like the processes, both halves that arise as a result are
fork()initially (almost) identical, but then they decide to do different things. As in unix, significant problems arise when cells continue to
forkgrow. Soon enough, they will exhaust resources, which can lead to death. This is called a tumor. The cells are stuffed with ulimitwatchdogs (watchdogs) to prevent the development of such a scenario. For example, the number of divisions is limited by shortening telomeres .A cell cannot clone itself until extremely stringent conditions are met - the default security settings . And only if all these defense mechanisms fail, a tumor may develop. As in computer security, it is difficult to find a balance between security (“no cells can share”) and usability.
You can compare this with a known problem with a stop , first described by computer science founder Alan Turing. It seems that to predict whether a program will ever end is just as easy as creating a functional genome that will never lead to cancer?
Mirroring, fault tolerance
 Each DNA helix is redundant in itself - it looks like a twisted ladder in which each step contains two bases - hence the word “pair of bases”. If one of these bases is missing, it can be removed from the base on the other side of the "step". T is always associated with A, C is always with G. So we can argue that the genome is mirrored inside the helix, like RAID-1.
Each DNA helix is redundant in itself - it looks like a twisted ladder in which each step contains two bases - hence the word “pair of bases”. If one of these bases is missing, it can be removed from the base on the other side of the "step". T is always associated with A, C is always with G. So we can argue that the genome is mirrored inside the helix, like RAID-1. Moreover, the chromosomes are always presented in duplicate - one from each parent, with one notable exception - the Y chromosome, which occurs only in males. Although the details are somewhat more complicated, we can assume that we have two copies of most genes. In the event that one is damaged or mutated into a useless state, we still have a second, independent copy. We usually call this failover.
Heap API, hell dependencies
During the interaction within the cell, proteins rely on the characteristics of each other. In 2001-2002, studies were published that proteins that interact with a large number of other proteins are not able to evolve, or at least do it very slowly (Nature Journal, June 28, 2001, and M. Kimura, T. Ohta in Science journal, April 26, 2002).
The authors of the research suggest that this is due to the huge number of internal dependencies that prevent a change in the protein “contract”. It was also noted that evolution is still happening, but very slowly, since both sides of the dependency must simultaneously evolve in a compatible way.
Viruses, worms
Recently, during the conversation, someone suggested that it would be really cool to break the genome and compromise by inserting a code that would copy itself to other genomes, using the host body as a transport. "Just like the Nimda worm!" He
soon realized that this is what biological viruses have been doing for millions of years. And they are very good at it.
A large number of such viruses have become an integral part of our genome and ride with all of us. To do this, they have to hide from the virus scanner, which seeks to detect someone else's, malicious code and prevent its penetration into the DNA.
Central dogma: .c -> .o -> a.out/.exe
At the dawn of the discoveries of the foundations of genetics, scientists had to deal with a variety of chemicals, the interrelationship of which was not obvious. The moment when it became clear from what it turns out, was proclaimed a great triumph and received the name "Central Dogma."
This dogma tells us that DNA is needed to get RNA, and RNA is needed to get proteins, just as it turns from an .c file into an .o object file, which can then be compiled into an executable file (a.out / exe) . In addition, she tells us that this is the only order in which the information flow follows.
And yet, the Central Dogma has recently faded a little. As with any billion-year project development, there were a lot of hacks, because of which information sometimes flows in the opposite direction. Sometimes RNA patches DNA, and it happens that DNA is modified by proteins called previously created.
But, in general, the dependencies are clear, so the Central Dogma still remains important.
Binary patches, aka "gene therapy"
Making fraud with DNA is pretty simple. There are companies to which you can send a file with DNA written in ASCII characters, and they synthesize relevant materials for you. We can even make up the DNA of the embryos of animals or plants.
It is much more difficult to patch an “executable file” right at runtime, and any programmer will confirm this. The same is with the genome. To change a running copy (of a person, for example), you need to edit every instance of the gene that you want to patch in the body.
For many years, medical science has been trying to patch people suffering from severe combined immunodeficiency (SCID, Severe Combined Immunodefeciency) - an extremely unpleasant disease that essentially disables the immune system, making the patient seriously ill. For a long time it became clear exactly what letters in the DNA must be corrected for the healing of these people.
Many attempts have been made to patch the “working” people with the help of viruses that would insert a new DNA into living organisms, but this approach has proved too complicated. The genome is extremely well protected to make such a simple approach — the cells store their code more reliably than Microsoft!
Nevertheless, a suitable virus was recently found that is able to break through the protection and correct DNA breaks, which, like,heals the sick .
Code Regression
As bugs are fixed in computer programs, we often introduce new bugs. The genome is replete with similar bugs. African Americans are immune to malaria, but instead are susceptible to sickle-cell anemia:
In tropical regions of the world where malaria is spread by parasites, people with one copy of a specific genetic mutation have an advantage in survival.
...
While inheriting one copy of this mutation is beneficial, inheriting two copies leads to tragedy. A child born with two copies of a genetic mutation has sickle cell anemia, a painful disease that affects red blood cells.
A source
And this is not the only example of such a regression.
Reed-Solomon codes: Forward error correction
Like a computer drive, DNA (and its intermediate form, RNA) can be damaged. To prevent common “one-bit errors”, protein coding in DNA symbols is redundant. There are 4 symbols of RNA: U, C, G, and A — in other words, a “byte” is 2 bits long. Three characters correspond to the amino acid.
Theoretically, 6 bits could correspond to 64 amino acids, however there are only 20 standard amino acids. For example, UCU, UCC, UCA and UCG - encode the same serine, whereas only UGG corresponds to tryptophan.
Then, it turns out that some of the most likely "typos" (UCU -> UCC) in the code will lead to the synthesis of the same amino acid. Read more about this phenomenon in the book " Metamagical Themas"by Douglas Hofstadter (Douglas Hofstadter).
Holy code: / * you are not required to understand this code * /
Some parts of the code are sacred. We may not remember who or why wrote it, we just know that it works. The guy who wrote it may have long left the company. With this code it is better not to get involved.
The concept of molecular hours is known in DNA. Some parts of the genome are actively changing, and some are inviolable. An excellent example of the latter is the histone genes H3 and H4.
These genes underlie how the genome is stored, and therefore are of paramount importance. Any failure in their code leads to a non-viable organism.
Therefore, we can assume that this code does not change in haste, and, apparently, the way it is. H3 and H4 genes have zeroeffective mutation rate in humans. But it goes even further. This code is common to everyone, starting with a person, ending with grass or mold.
The rate of nucleotide substitution in terms of a site for 1 billion years between various protein-coding genes of humans and rodents with a spread of 80 million years, based on fossil evidence:Gene Number of Codons Effective tempo Histon 3 135 0.00 Histone 4 101 0.00 Insulin 51 0.13 Gamma Interferon 136 2.79
A source
Apparently, there are two ways by which the genome can verify that there is no code mutation. The first method was described earlier: to use amino acids, the coding of which is very redundant, which will allow to get the same result even in the case of "typos".
The second way is that the genes can be copied at the beginning or end of the cell's reproduction process, which provides more or less favorable conditions for copying. In addition, a huge number of similar conditions are used.
It seems that H3 and H4 were very carefully worked out, since they allow a lot of "synonymous" changes, which, thanks to the clever techniques described above, do not lead to a change in the result.
Framing errors: start and stop bits
...0 0000 0001 0000 0010 0000 0011 0...This sequence obviously describes the 8-bit values 1, 2, and 3. I added spaces to show where the byte begins and ends. Many serial devices use stop and start bits to indicate where you need to start reading. If we move this sequence a little:
...00 0000 0010 0000 0100 0000 0110 ...it will suddenly begin to denote 2, 4, 6! To avoid this, signals are used in the DNA that are generated that tell the cell where to start reading. It is curious that there are parts of the genome that you can start reading from several starting points, and each time get a useful (but different) result. That's what I call a cool hack!
Each way of reading such a piece of DNA is called an open reading frame and there are usually 6 of them, 3 in each direction.
Mass multiprocessing: every cell is a universe
And yet, DNA is not like a programming language. She really is not. However, there are some big similarities. You can consider each cell as a central processor, each of which runs its own OS kernel. Each cell has a complete copy of the core, but only activates the corresponding parts - its own modules and drivers, so to speak.
When a cell needs to do something (“call a function”), it finds the necessary part of the genome and transcribes it into RNA. The RNA is then translated into a sequence of amino acids, which together give the protein that was encoded in the DNA. And now the coolest part!
This protein will be labeled with a shipping address. It is presented in the form of a marker consisting of several amino acids, which tells the cell exactly where to deliver this protein. Inside the cell there is a whole mechanism that carries out these instructions and is able to deliver the protein to an address that may even be outside the cell itself.
After delivery, this instruction comes off and several post-processing steps are carried out, which can, for example, activate protein, because transporting active protein through places for which it is not intended is not the best idea.
Makefile
Organisms usually begin their existence from a single cell, which, as mentioned earlier, contains two full-fledged copies of the genome. A peculiar tar file in which all files are already unpacked, ready to use. What next?
Meet the homeobox. Cells must be copied and each must be assigned its purpose of existence. Genes that contain a homeobox begin to build a “top-down” dependency chain, which means “start from the head.” In order for this to work, a chemical gradient is formed, thanks to which the cells can sense where they are and determine their further actions required for the formation of the head or for the formation of the notochord.
First discovered in 1983, homeobox genes are currently one of the most exciting areas to explore. Note the curious fact that, like the makefile, the “HOX” genes are only responsible for the inclusion of different things in other genes and in themselves form nothing.
It seems that the “syntax” of the homeobox is “sacred” in the sense described above. What happens if you copy the “HOX” part of the mouse gene, which is the “leg selector” to the Drosophila homeoobox?
In fact, in the case of inserting the HOX-B6 mouse gene into the Drosophila genome, it can replace the antennapedia genes and grow legs on the antenna site.
A source
The genomes of fruit flies and humans branched not millions but hundreds of millions of years ago. And yet you can copy the plots (“selectors” in the language of genetics) makefiles, and they will work. Please note that the “growing legs” procedure in a fruit fly is completely different from a mouse, but the “selector” is able to correctly run the necessary instructions.
Plugins: Plasmids
All living organisms have DNA, sometimes organized into several chromosomes (“libraries”), sometimes only one (usually looped out in such cases). This also applies to most bacteria. And in the neighborhood of this large main genome, such bacteria often contain “plasmids” - tiny DNA rings with specific functions.
These plasmids are partly compatible with different species and are transmitted horizontally using a variety of mechanisms. In this way, for example, even non-identical bacteria can “learn” antibiotic resistance from each other.
When compared to the world of programming, plasmids are not a voluntary acquisition, and are similar to loading .so using LD_PRELOAD or its equivalent on other platforms. Indeed, plasmids are often injected for research purposes. They can be introduced into any kind of bacteria and immediately get to work.
Plasmids copy themselves independently of the main chromosome, therefore they become a constant component of the bacterium. To achieve this, a plasmid possesses a gene with the great name “replication origin”, which is triggered when a cell is about to divide.
