Expansion of the front of bacteria in the arena with antibiotics: a spectacular experiment at Harvard Medical School
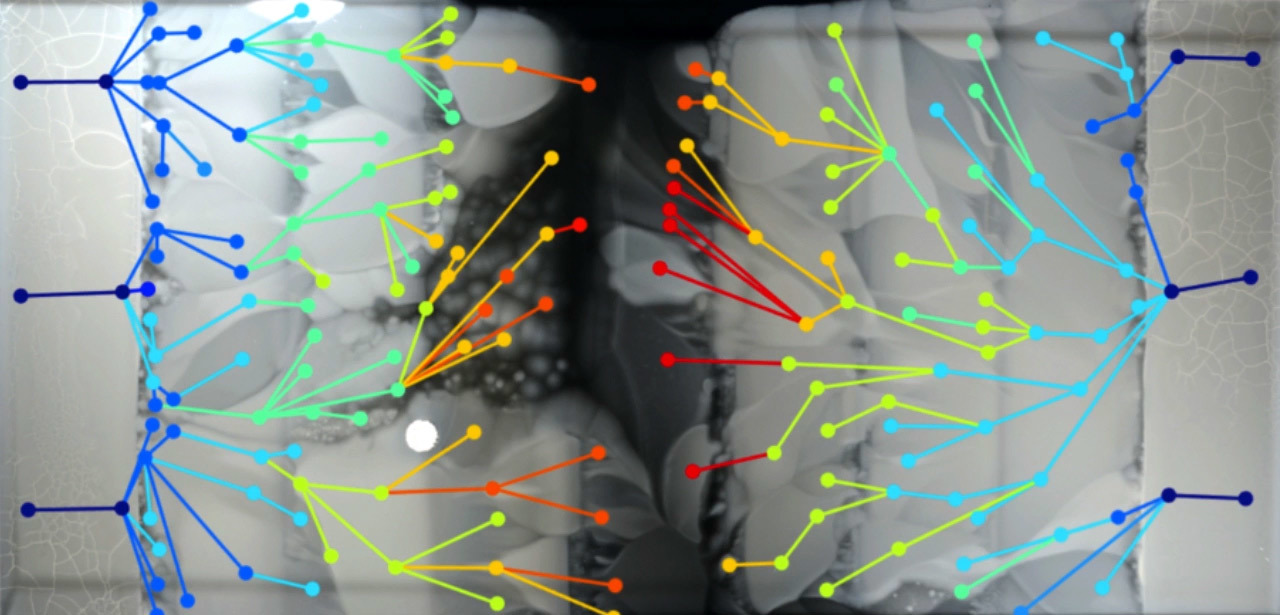
As the resistance from the antibiotic increases in the bacterial population, parallel lines of evolution appear, which differ in phenotype and genotype. Shot from Harvard Medical School demo video
Usually in laboratory experiments bacteria are bred in a homogeneous environment. Scientists from Harvard Medical School went further. They organized an unusual experiment on the evolution of bacteria in a mixed environment - on a huge “platter” measuring 120 × 60 cm.
The space was divided into zones with different concentrations of the antibiotic, so that only generations of bacteria with suitable mutations pass into the next zone. As a result, in the final round in the center of the arena, “supermicrobes” make their way through, that is, they maximally change their genotype and phenotype as a result of evolutionary selection (see the isolation of the opportunists at the end of the article).
The microbial battlefield of antibiotics is called MEGA (microbial evolution and growth arena). Such an environment is specially created so that with an exponential growth in the number of bacteria they do not compete with each other for limited resources, as is customary in most scientific experiments. Here, the resources are practically unlimited, and from bacteria it is only necessary to capture new territories and adapt to new conditions of life, multiplying their numbers with almost no restrictions. In this sense, the movement of the bacterial front resembles the expansion of the human species on the planet Earth in the Middle Ages with the colonization of new territories (presumably, the expansive development of the human race will continue beyond the boundaries of the home planet with the colonization of all habitable territories within reach.)
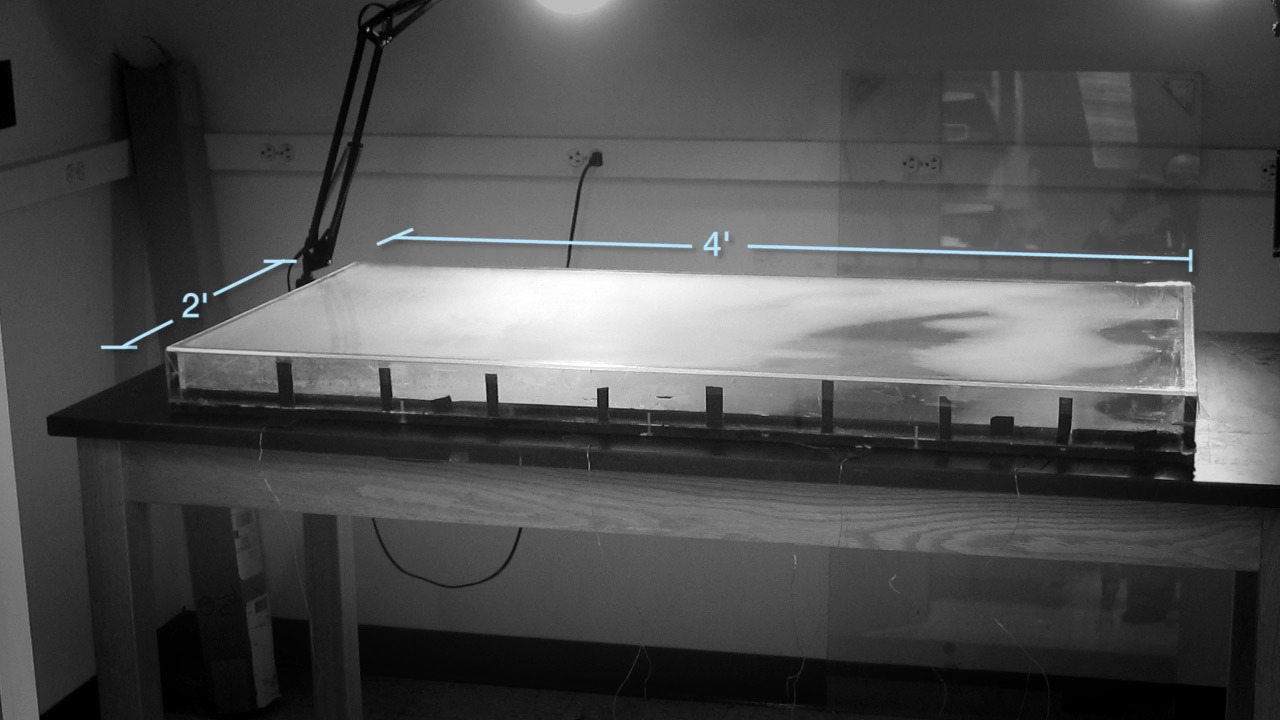
Arena MEGA.Shot from Harvard Medical School demo video The
experiment has not only scientific, but also educational value. The large space of the MEGA arena allows you to visually observe mutations and natural selection as the front of the bacterial population spreads. An impressive sight.
Earlier studies have shown that structured microenvironments of this type increase the rate of evolution in small populations of bacteria with a change in genotype ( Q. Zhang et al., Science 333, 1764–1767 (2011) ). But until now, the question how exactly this happens in large populations has remained unexplored.
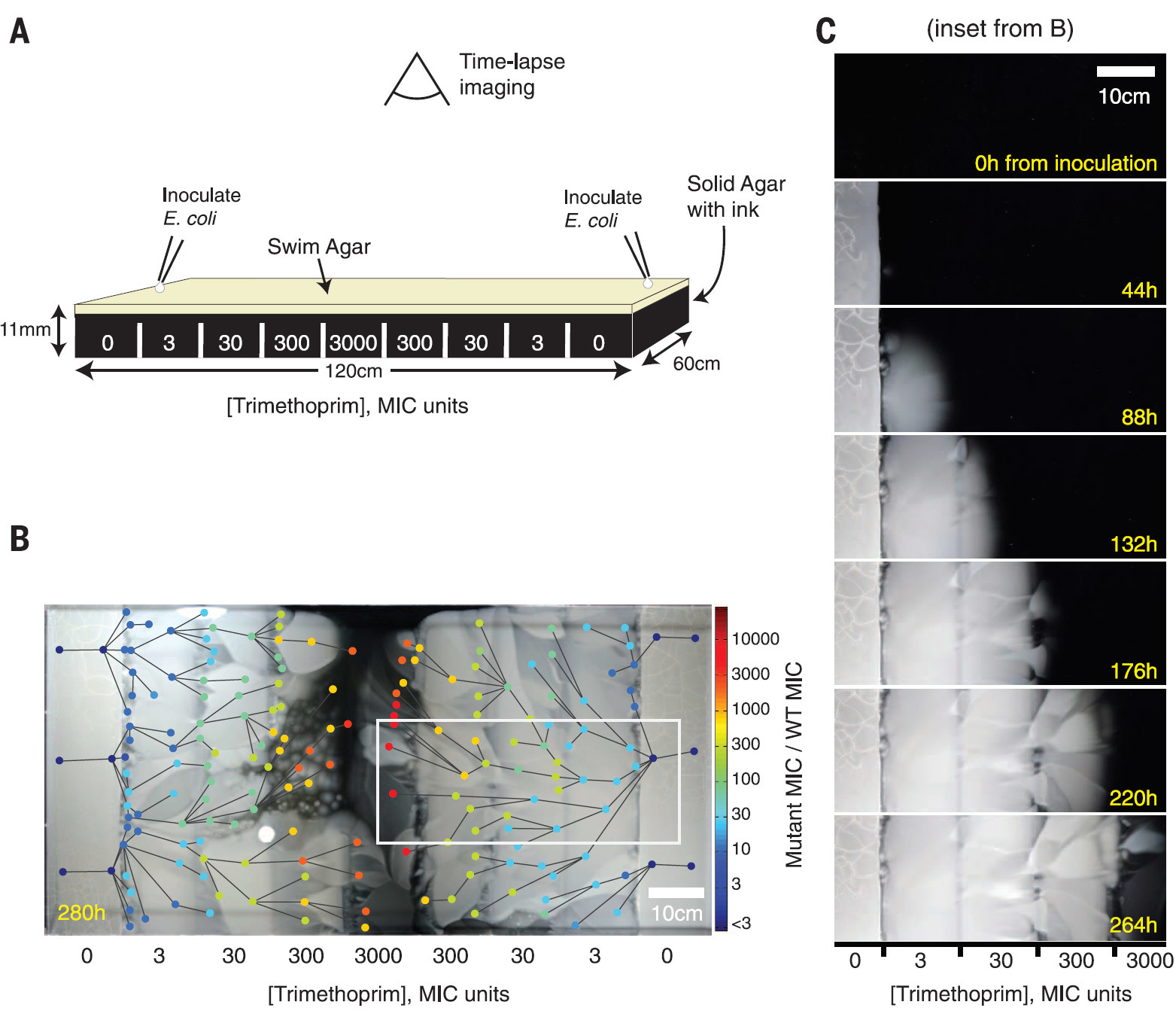
For such an experiment, a rectangular Petri dish measuring 120 × 60 cm was designed, structured into zones with an exponential increase in the concentration of antibiotic trimethoprim from the periphery to the center, as well as with nutrients for the reproduction of bacteria. A large area of the arena did not allow E. coli bacteria to mix with each other in order to more clearly observe the mutations occurring.
The construction of the MEGA arena and the result of the propagation of the bacterial front for 12 days are shown in illustration B. Circles of different colors indicate 182 types of mutated bacteria, the color indicates the concentration of bacteria. The lines between the species correspond to the direction of mutation, based on the video data.
As the antibiotic continues to increase in the bacterial population, numerous parallel lines of evolution emerge that differ in phenotype and genotype.
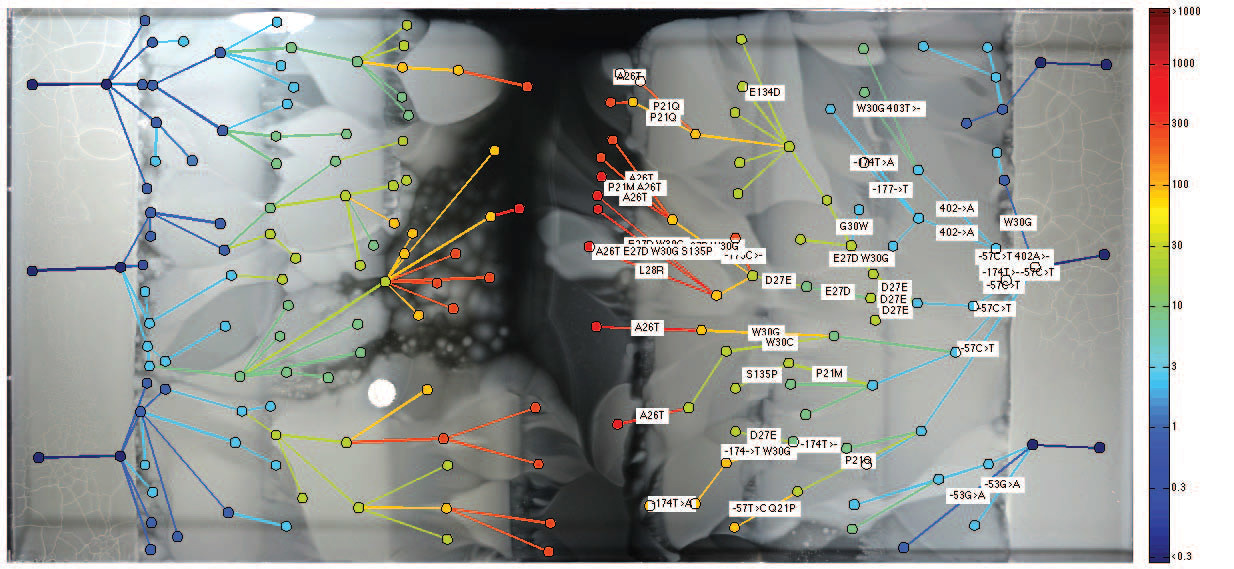
After examining the bacteria in the forefront and behind the front of the bacterial population, scientists found out some interesting things. It turned out that evolution is not always driven by bacteria that are most resistant to antibiotics. Strangely enough, sometimes the most stable hereditary lines are locked behind more sensitive bacteria. Apparently, this is due to "premature" mutations, when some bacteria are ready to survive in a higher concentration of antibiotic, which will appear in the future, but has not yet appeared. In such a situation, potentially more adapted bacteria give place in the front to their relatives, who are adapted precisely to the actual concentration that exists at the moment.
To test this theory, scientists took samples of isolated bacterial colonies with “premature” mutations and forcibly placed them in front of the front. As expected, they survived in conditions in which the main bacterial front could not survive.

The spatial trap of compensatory mutations are bacteria that are so ahead of their time that even after the occurrence of suitable conditions, they are already locked behind the advancing front. Illustration: Harvard Medical School
Scientists have carefully studied the genotype of the most mutated species of bacteria that managed to survive in the solution with the maximum concentration of trimethoprim. It turned out that in these species, the folA gene, which encodes dihydrofolate reductase (DHFR) and is the target of trimethoprim, has most often been mutated. The greater the bacterium's resistance to the antibiotic, the greater the mutations in this gene. In addition, mutations were found in several other genes unrelated to the action of a particular antibiotic. Among them were the mar and sox operons , which are responsible for the reaction to stress. It was previously known that these "stressful" genes play an important role for successful resistance to antibiotics.
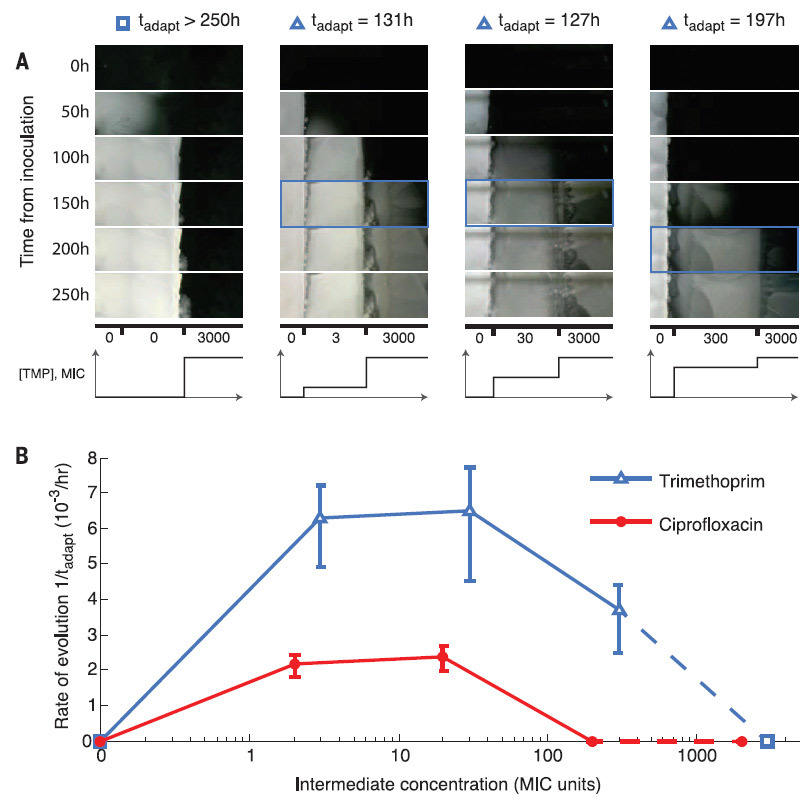
Scientists also found that better adaptation to the weak effects of the antibiotic speeds up later adaptation to higher concentrations (in the illustration below). Just like people who are able to better adapt to deteriorating living conditions, if changes occur gradually and imperceptibly.
The experiment is described in a scientific article published September 9, 2016 in the journal Science (doi: 10.1126 / science.aag0822).
