AlphaFold: Using AI for Scientific Discoveries
- Transfer
Hello again! We are sharing the publication, the translation of which was prepared especially for students of the course "Neural Networks in Python" .

Today we will talk about the first important event in the history of DeepMind to show how research using artificial intelligence can stimulate the emergence of scientific discoveries. Due to the interdisciplinary nature of our work, DeepMind brought together experts from the fields of structural biology, physics and machine learning to use advanced methods for predicting the three-dimensional structure of a protein based solely on its genetic sequence.
The AlphaFold system that we have been working on over the past two years is based on many years of research experience using extensive genome data to predict protein structure. The three-dimensional protein models that AlphaFold generates are much more accurate than those obtained previously. This marked significant progress in one of the main tasks of biology.
Proteins are large and complex molecules needed to sustain life. Almost all the functions of our body, whether it is muscle contraction, light perception, or the conversion of food into energy, can be traced to one or more proteins and how they move and change. Recipes for these proteins, called genes, are encoded in our DNA.
The properties of a protein depend on its unique three-dimensional structure. For example, the antibody proteins that make up our immune system are “Y-shaped” and look like special hooks. Clinging to viruses and bacteria, antibody proteins are able to detect and label pathogens for subsequent destruction. Similarly, collagen proteins are in the form of cords that transmit tension between cartilage, ligaments, bones and skin. Other types of proteins include Cas9, which, guided by CRISPR sequences, act as scissors that cut DNA and insert new sites. Antifreeze proteins, whose three-dimensional structure allows them to bind to ice crystals and prevent freezing of organisms; and ribosomes, which act as a programmed conveyor that is involved in the construction of proteins.
Determining the three-dimensional structure of a protein solely from its genetic sequence is a complex task that scientists have been struggling with for decades. The problem is that DNA contains only information about the sequence of building blocks of a protein called amino acid residues that form long chains. Predicting how these chains will form a complex 3D protein structure is known as the “protein folding problem.”
The larger the protein, the more difficult it is to model, since more bonds are formed between amino acids that need to be taken into account. As follows from the Levintal paradoxTo list all the possible configurations of an ordinary protein, before its regular three-dimensional structure is reached, it will take more time than the Universe exists.

The ability to predict the shape of a protein is extremely useful because it is fundamental to understanding the role of protein in the body, as well as the diagnosis and treatment of diseases such as Alzheimer 's , Parkinson's , Huntington 's disease and cystic fibrosis , which doctors believe are caused by improperly folded proteins.
We are especially pleased that the ability to predict the shape of a protein can improve understanding of how our body works, this will allow us to develop new drugs efficiently. As we get more information about the forms of proteins and how they work through modeling, new opportunities for creating drugs open up and the cost of experiments decreases. Ultimately, these discoveries will improve the quality of life for millions of patients worldwide.
Understanding the process of protein folding can also help in developing a type of protein that will make a significant contribution to the surrounding reality. For example, advances made through the development of protein in biodegradable enzymes can help deal with contaminants such as plastic and oil, helping break down waste without damaging the environment. In fact, researchers have already begun to design bacteria that secrete proteins that make waste biodegradable and make it easier to handle.
In order to stimulate research and evaluate progress in the field of the latest methods for increasing the accuracy of forecasting, in 1994 a large-scale two-year competition was launched called the “Community Experiment on Critical Evaluation of Protein Structure Prediction Methods” (CASP), which has become the gold standard in valuation techniques.
Over the past five decades, scientists have been able to recognize the forms of proteins in the laboratory using experimental methods such as cryoelectronic microscopy , nuclear magnetic resonance, or X-ray diffraction , but each method has been deduced by many trials and errors that took years and cost tens of thousands of dollars. That's why biologists are now turning to AI methods as an alternative to the long and laborious process of researching complex proteins.
Fortunately, the field of genomics has enough data due to the rapid reduction in the cost of genetic sequencing. As a result, in the last few years, approaches have become increasingly popular.to the prediction problem using deep learning and based on genome data. DeepMind's work on this issue led to the appearance of AlphaFold, which we introduced to CASP this year. We are proud to be part of the progress that CASP experts have called “unprecedented progress in the ability of computational methods to predict the structure of a protein.” As a result, we took first place in the ranking of teams (we are A7D).
Our team focused precisely on the task of modeling target forms from scratch, without using previously solved proteins as templates. We achieved a high degree of accuracy in predicting the physical properties of the protein structure, and then used two different methods to predict complete protein structures.
Both of these methods used deep neural networks that are trained to predict the properties of a protein by its genetic sequence. The properties that the network predicts are: (a) the distance between pairs of amino acids and (b) the angles between the chemical bonds that connect these amino acids. The first development was a real progress in the use of popular methods that determine whether pairs of amino acids are next to each other.
We trained the neural network to predict a separate distribution of distances between each pair of protein residues. These probabilities were then combined into a score that shows how well designed the protein structure is. We also trained another neural network that uses all distances together to evaluate how close the proposed structure is to the correct answer.

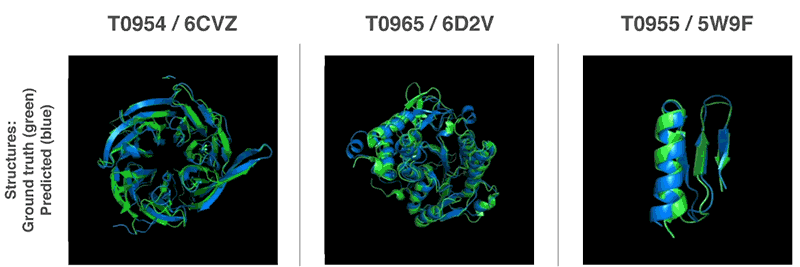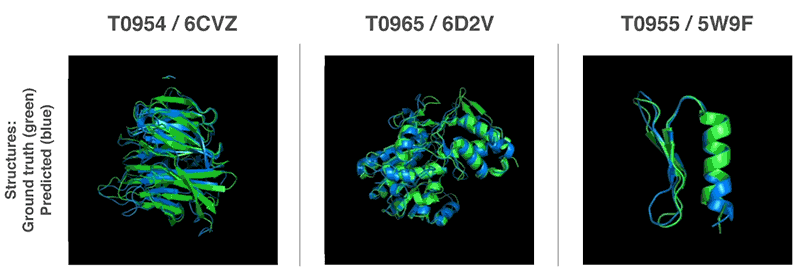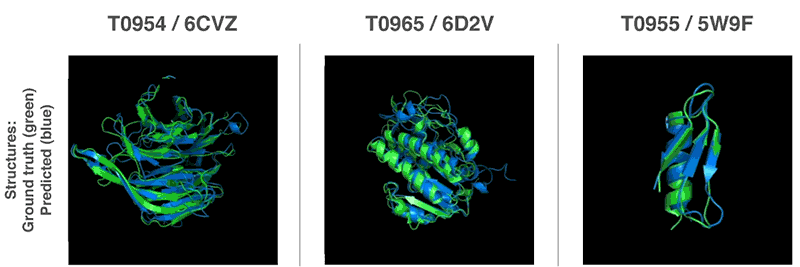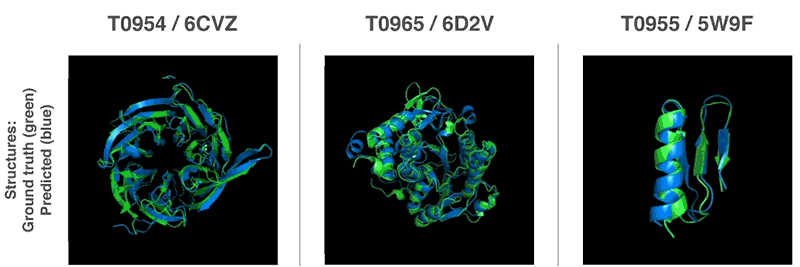
Using these valuation functions, we were able to find structures that match our forecasts. Our first method is based on methods widely used in structural biology; it has repeatedly replaced parts of the protein structure with new fragments. We trained the generative-competitive neural network to propose new fragments that are used to continuously improve the assessment of the proposed protein structure.

The second method optimized grades using gradient descent (the mathematical method commonly used in machine learning for small incremental improvements), which led to high accuracy of the structures. This method was applied to whole protein chains, rather than to pieces that must be stacked separately before assembly, which reduces the complexity of the prediction process.
The success of our protein coagulation pen test shows that machine learning systems can integrate a variety of information sources to help scientists quickly develop creative solutions to complex problems. We have already seen how AI helps people master complex games through systems such as AlphaGo and AlphaZero , we also hope that once the breakthrough of AI helps humanity solve fundamental scientific problems.
It is interesting to see the first progress in protein folding, demonstrating the usefulness of AI in making scientific discoveries. Even though we still have a lot to do, we clearly understand that we will be able to contribute to the search for treatment of various diseases, help the environment and much more, because in fact the potential is huge. With a dedicated team focused on exploring how machine learning can advance the world of science, we are exploring various ways and methods with which our technology can influence the world around us.

Today we will talk about the first important event in the history of DeepMind to show how research using artificial intelligence can stimulate the emergence of scientific discoveries. Due to the interdisciplinary nature of our work, DeepMind brought together experts from the fields of structural biology, physics and machine learning to use advanced methods for predicting the three-dimensional structure of a protein based solely on its genetic sequence.
The AlphaFold system that we have been working on over the past two years is based on many years of research experience using extensive genome data to predict protein structure. The three-dimensional protein models that AlphaFold generates are much more accurate than those obtained previously. This marked significant progress in one of the main tasks of biology.
What is the problem of protein folding?
Proteins are large and complex molecules needed to sustain life. Almost all the functions of our body, whether it is muscle contraction, light perception, or the conversion of food into energy, can be traced to one or more proteins and how they move and change. Recipes for these proteins, called genes, are encoded in our DNA.
The properties of a protein depend on its unique three-dimensional structure. For example, the antibody proteins that make up our immune system are “Y-shaped” and look like special hooks. Clinging to viruses and bacteria, antibody proteins are able to detect and label pathogens for subsequent destruction. Similarly, collagen proteins are in the form of cords that transmit tension between cartilage, ligaments, bones and skin. Other types of proteins include Cas9, which, guided by CRISPR sequences, act as scissors that cut DNA and insert new sites. Antifreeze proteins, whose three-dimensional structure allows them to bind to ice crystals and prevent freezing of organisms; and ribosomes, which act as a programmed conveyor that is involved in the construction of proteins.
Determining the three-dimensional structure of a protein solely from its genetic sequence is a complex task that scientists have been struggling with for decades. The problem is that DNA contains only information about the sequence of building blocks of a protein called amino acid residues that form long chains. Predicting how these chains will form a complex 3D protein structure is known as the “protein folding problem.”
The larger the protein, the more difficult it is to model, since more bonds are formed between amino acids that need to be taken into account. As follows from the Levintal paradoxTo list all the possible configurations of an ordinary protein, before its regular three-dimensional structure is reached, it will take more time than the Universe exists.

Why is protein folding important?
The ability to predict the shape of a protein is extremely useful because it is fundamental to understanding the role of protein in the body, as well as the diagnosis and treatment of diseases such as Alzheimer 's , Parkinson's , Huntington 's disease and cystic fibrosis , which doctors believe are caused by improperly folded proteins.
We are especially pleased that the ability to predict the shape of a protein can improve understanding of how our body works, this will allow us to develop new drugs efficiently. As we get more information about the forms of proteins and how they work through modeling, new opportunities for creating drugs open up and the cost of experiments decreases. Ultimately, these discoveries will improve the quality of life for millions of patients worldwide.
Understanding the process of protein folding can also help in developing a type of protein that will make a significant contribution to the surrounding reality. For example, advances made through the development of protein in biodegradable enzymes can help deal with contaminants such as plastic and oil, helping break down waste without damaging the environment. In fact, researchers have already begun to design bacteria that secrete proteins that make waste biodegradable and make it easier to handle.
In order to stimulate research and evaluate progress in the field of the latest methods for increasing the accuracy of forecasting, in 1994 a large-scale two-year competition was launched called the “Community Experiment on Critical Evaluation of Protein Structure Prediction Methods” (CASP), which has become the gold standard in valuation techniques.
How will AI make a difference?
Over the past five decades, scientists have been able to recognize the forms of proteins in the laboratory using experimental methods such as cryoelectronic microscopy , nuclear magnetic resonance, or X-ray diffraction , but each method has been deduced by many trials and errors that took years and cost tens of thousands of dollars. That's why biologists are now turning to AI methods as an alternative to the long and laborious process of researching complex proteins.
Fortunately, the field of genomics has enough data due to the rapid reduction in the cost of genetic sequencing. As a result, in the last few years, approaches have become increasingly popular.to the prediction problem using deep learning and based on genome data. DeepMind's work on this issue led to the appearance of AlphaFold, which we introduced to CASP this year. We are proud to be part of the progress that CASP experts have called “unprecedented progress in the ability of computational methods to predict the structure of a protein.” As a result, we took first place in the ranking of teams (we are A7D).
Our team focused precisely on the task of modeling target forms from scratch, without using previously solved proteins as templates. We achieved a high degree of accuracy in predicting the physical properties of the protein structure, and then used two different methods to predict complete protein structures.
Using neural networks to predict physical properties
Both of these methods used deep neural networks that are trained to predict the properties of a protein by its genetic sequence. The properties that the network predicts are: (a) the distance between pairs of amino acids and (b) the angles between the chemical bonds that connect these amino acids. The first development was a real progress in the use of popular methods that determine whether pairs of amino acids are next to each other.
We trained the neural network to predict a separate distribution of distances between each pair of protein residues. These probabilities were then combined into a score that shows how well designed the protein structure is. We also trained another neural network that uses all distances together to evaluate how close the proposed structure is to the correct answer.


New methods for predicting protein structures
Using these valuation functions, we were able to find structures that match our forecasts. Our first method is based on methods widely used in structural biology; it has repeatedly replaced parts of the protein structure with new fragments. We trained the generative-competitive neural network to propose new fragments that are used to continuously improve the assessment of the proposed protein structure.

The second method optimized grades using gradient descent (the mathematical method commonly used in machine learning for small incremental improvements), which led to high accuracy of the structures. This method was applied to whole protein chains, rather than to pieces that must be stacked separately before assembly, which reduces the complexity of the prediction process.
What's next?
The success of our protein coagulation pen test shows that machine learning systems can integrate a variety of information sources to help scientists quickly develop creative solutions to complex problems. We have already seen how AI helps people master complex games through systems such as AlphaGo and AlphaZero , we also hope that once the breakthrough of AI helps humanity solve fundamental scientific problems.
It is interesting to see the first progress in protein folding, demonstrating the usefulness of AI in making scientific discoveries. Even though we still have a lot to do, we clearly understand that we will be able to contribute to the search for treatment of various diseases, help the environment and much more, because in fact the potential is huge. With a dedicated team focused on exploring how machine learning can advance the world of science, we are exploring various ways and methods with which our technology can influence the world around us.
