Swiss json processing knife
How to work effectively with json in R?
It is a continuation of previous publications .
Formulation of the problem
As a rule, the main source of data in json format is the REST API. The use of json, in addition to platform independence and the convenience of human perception of data, allows exchanging unstructured data systems with a complex tree structure.
In the tasks of building an API, this is very convenient. It is easy to ensure the versioning of communication protocols, it is easy to provide the flexibility of information exchange. At the same time, the complexity of the data structure (nesting levels can be 5, 6, 10 or even more) is not scary, because writing a flexible parser for a single record that takes into account everything and everything is not so difficult.
Data processing tasks also include obtaining data from external sources, including in json format. R has a good set of packages, in particular, jsonliteintended for converting json to R objects ( listor data.frame, if the data structure allows).
However, in practice, two classes of problems often arise, when the use of jsonlitesimilar ones becomes extremely ineffective. Tasks look something like this:
- processing a large amount of data (unit of measure - gigabytes) obtained during the operation of various information systems;
- combining a large number of variable-structured responses received during a packet of parameterized REST API requests into a uniform rectangular representation (
data.frame).
An example of a similar structure in the illustrations:
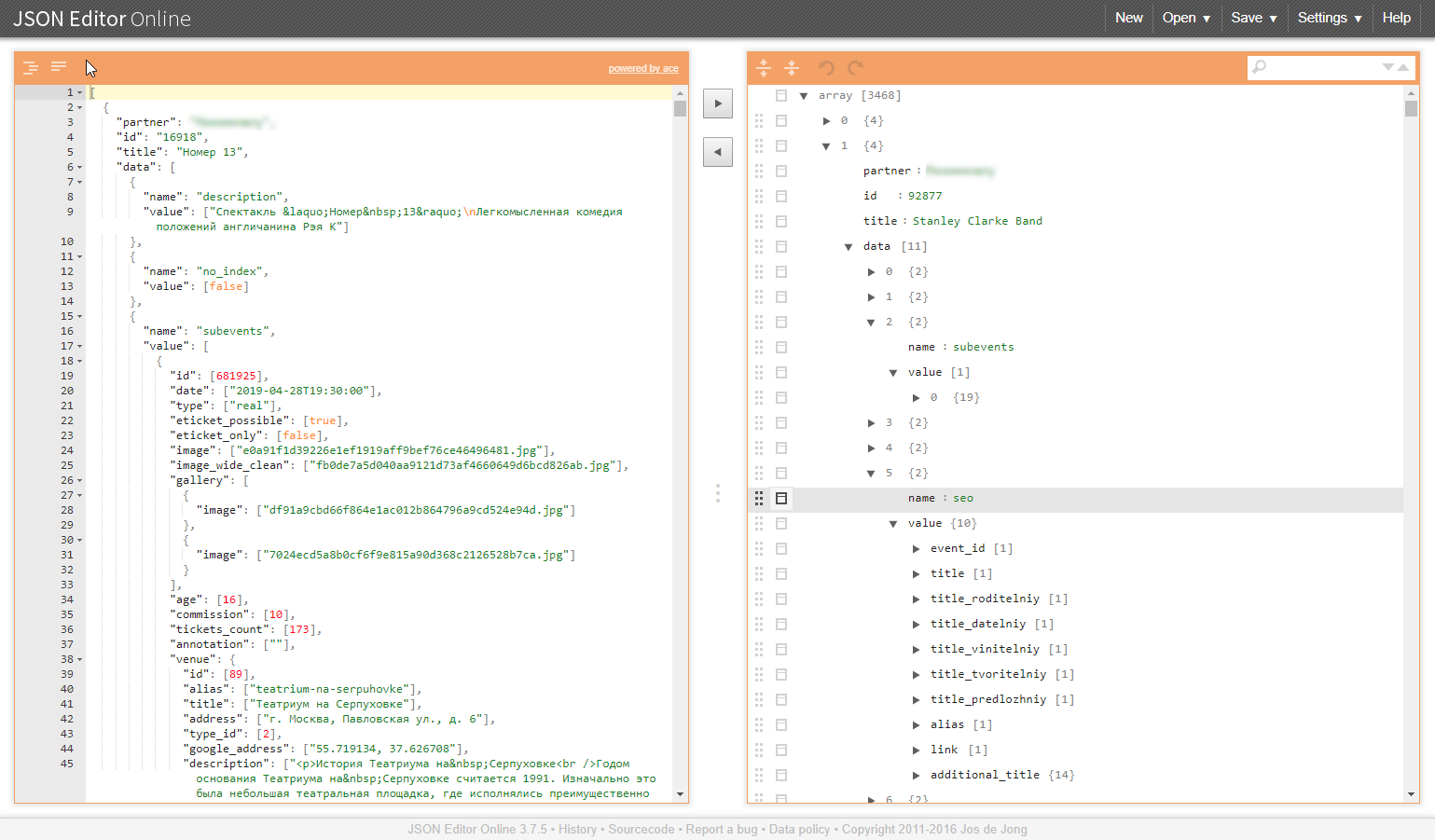
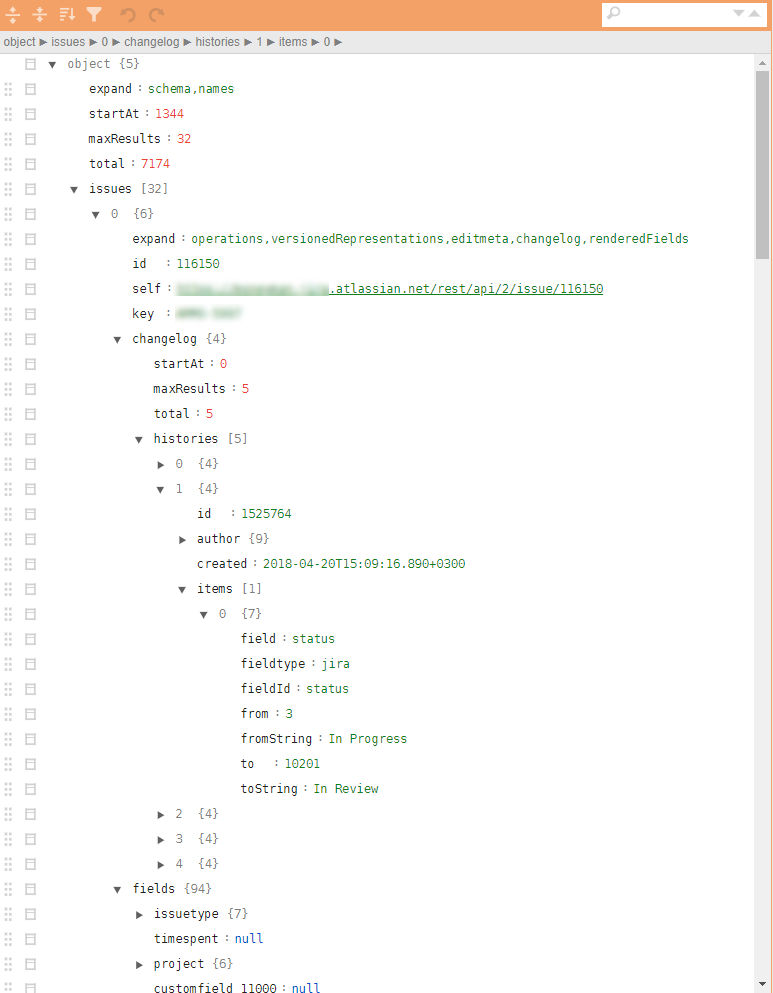
Why are these task classes problematic?
Large amount of data
As a rule, unloading from information systems in json format is an indivisible data block. To parse it correctly, you need to read it all and go over its entire volume.
Induced problems:
- a corresponding amount of RAM and computing resources is needed;
- parsing speed strongly depends on the quality of the libraries used, and even if there are sufficient resources, the conversion time can be tens or even hundreds of minutes;
- in the event of a parsing failure, no result is obtained at the output, and there is no reason to hope that everything will always go smoothly, there is no reason, rather the opposite;
- It will be very successful if the parsed data can be converted to
data.frame.
Merging Tree Structures
Similar tasks arise, for example, when it is necessary to collect the directories required by the business process for work by a packet of requests through the API. In addition, directories imply unification and readiness for embedding into the analytical pipeline and potential uploading to the database. And this again makes it necessary to turn such summary data into data.frame.
Induced problems:
- tree structures themselves will not turn into flat ones. json parsers turn the input data into a set of nested lists, which then manually need to be deployed for a long time and painfully;
- freedom in attributes of the data output (idle or may not be granted) leads to the appearance of
NULLobjects which are relevant in the lists but can not "fit" indata.framewhich further complicates the postprocessing and complicates even basic process of merging the individual lines-sheetsdata.frame(whetherrbindlist,bind_rows, 'map_dfr' orrbind).
JQ - way out
In particularly difficult situations, the use of very convenient approaches of the package jsonlite“convert everything to R objects” for the above reasons gives a serious malfunction. Well, if you manage to get to the end of processing. Worse, if in the middle you have to spread your arms and give up.
An alternative to this approach is to use the json preprocessor, which operates directly on json data. Library jqand wrapper jqr. Practice shows that it is not only used little, but few have heard of it at all, and in vain.
The benefits of the library jq.
- the library can be used in R, in Python and on the command line;
- all transformations are performed at the json level, without transformation into representations of R / Python objects;
- processing can be divided into atomic operations and use the principle of chains (pipe);
- cycles for processing object vectors are hidden inside the parser, the iteration syntax is maximally simplified;
- the ability to carry out all procedures for the unification of the json structure, the deployment and selection of the necessary elements in order to form a json format that can be converted in batches into
data.frametoolsjsonlite; - multiple reduction of R code responsible for processing json data;
- huge processing speed, depending on the volume and complexity of the data structure, the gain can be 1-3 orders of magnitude;
- much less RAM requirements.
The processing code is compressed to fit the screen and may look something like this:
cont <- httr::content(r3, as = "text", encoding = "UTF-8")
m <- cont %>%
# исключим ненужные атрибутивные списки
jqr::jq('del(.[].movie.rating, .[].movie.genres, .[].movie.trailers)') %>%
jqr::jq('del(.[].movie.countries, .[].movie.images)') %>%
# исключим ненужные атрибутивные списки
jqr::jq('del(.[].schedules[].hall, .[].schedules[].language, .[].schedules[].subtitle)') %>%
# исключим ненужные атрибутивные списки
jqr::jq('del(.[].cinema.location, .[].cinema.photo, .[].cinema.phones)') %>%
jqr::jq('del(.[].cinema.goodies, .[].cinema.subway_stations)')
# расщепляем
m2 <- m %>%
jqr::jq('[.[] | {date, movie, schedule: .schedules[], cinema}]')
df <- fromJSON(m2) %>%
as_tibble()jq is very elegant and fast! To those to whom it is relevant: download, set, understand. We speed up processing, we simplify life for ourselves and our colleagues.
Previous post - “How to start applying R in Enterprise. An example of a practical approach . ”
