Tic-tac-toe: demonstration of the controlled process of DNA structure reconfiguration

Scientists held a batch of tic-tac-toe with DNA. It sounds funny, but this is only the tip of the iceberg diminished by a slight humor from the scientists themselves. In fact, today's research reveals new methods for controlling the kinetics of the interaction of DNA-nanostructures, which together can perform complex tasks. Using DNA as the main part of the system is not an easy task, but this does not stop scientists, whose minds are full of ideas, but hearts of enthusiasm. And so, how did they manage to move the DNA blocks according to a given pattern, what did the research show and what are the prospects for this technique in the future? For answers, we turn to the research group report. Go.
The basis of the study
In this study, scientists have demonstrated a new method of moving DNA tiles, which is based on the principles of the reference connection and the branch offset. These principles are similar to the standard DNA strand bias, but are aimed at larger objects, more precisely, DNA structures.
This technique allowed us to gain control over the kinetics of the DNA of the plates and to realize the desired chain of reactions within the multi-structural systems. The game of tic-tac-toe was not held for fun, but to demonstrate the possibility of shifting the DNA of the tile in any order and position.
To understand the mechanisms of biological systems, it is necessary to consider its main components, among which the most important are molecular structures. DNA, as an information carrier molecule, is considered one of the best materials for studying and creating self-organized nanostructures.
Among the techniques that are based on DNA, DNA origami has become more and more popular in recent years, allowing us to create DNA structures of any desired shape and configuration.
The constituent elements of one DNA origami can be twisted — twisted as you please, but it has not yet been possible to achieve controlled interaction between several DNA origami, as constituent elements of a larger, and therefore more complex, structure. At the moment, you could only connect / disconnect several DNA origami.
In today's study, scientists have demonstrated a new method of DNA origami interaction. There is a structure in which a DNA origami tile is inserted, displacing another tile from the common array and connecting to the structure at the expense of the binding domain * at the edge of the tile (support / bond point).
Binding / binding domain * is a domain of a protein that is part of the protein chain, but is able to function separately from it.As an example, scientists have created three options for reconfiguration: competitive, consistent and cooperative. The combination of these “movements” (step-by-step reconfiguration) was demonstrated as a tic-tac-toe game when each player had 9 unique DNA origami that can be moved in any order to a field at 264x264 nm.
Formation of DNA tiles
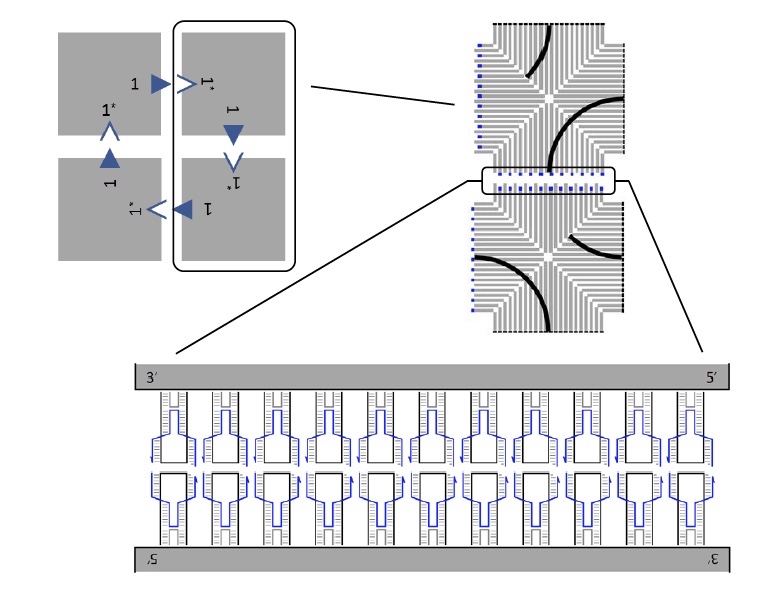
Structure of DNA tiles.
Earlier, before conducting this study, scientists have developed a method for creating individual DNA tiles. Each tile on the two edges has 11 brackets, each of which participates in the stacking.
The image above shows an array of 4 tiles (top left), each of which is connected to the next by means of brackets.
Researchers considered several possible options for creating larger arrays with a large number of tiles. For this, in theory, it was necessary that the bond between the tiles was rather weak. This would allow it to break and rebuild the array. However, observations have shown that it is possible to create reconfigurable multi-array arrays even at temperatures when the connection between tiles must be permanent.
Reconfig tiles.
This phenomenon has two possible explanations. The first is that the formation of bonds occurs at a sufficiently high temperature, while falling within the temperature range of the reversibility of the process. Secondly, the dimer and trimer undergo a displacement reaction, therefore a 2x2 array is formed when the monomer is released. They can also make 2 copies of trimers, forming a 2x2 array, releasing the dimer.
If during the displacement reactions there is no spontaneous detachment of the tiles, then there will be no kinetic traps, due to which the tiles will be assembled into a 2x2 configuration.
It was necessary to check whether one DNA origami would crowd out another in the general structure, while not spontaneously linking within the structure itself. In other words, whether DNA origami will be amateur or not. For this, two experiments were carried out.
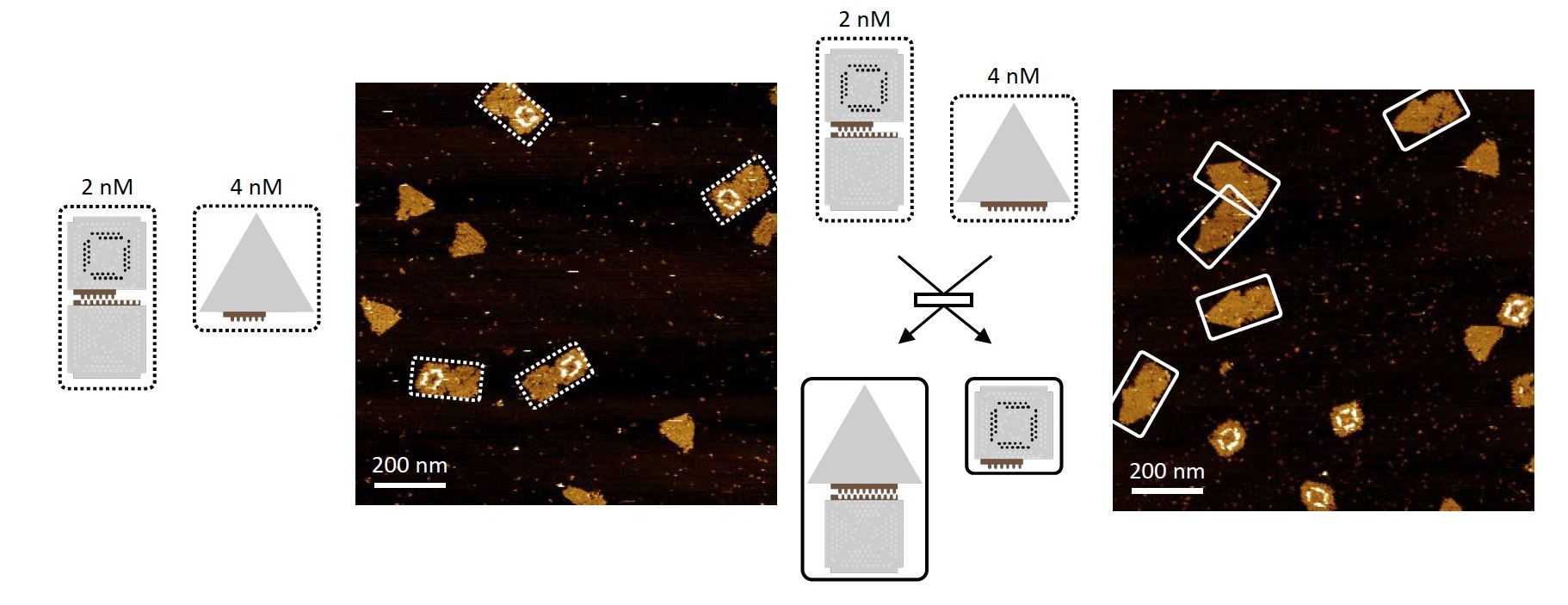
Practical experience: 2 DNA tiles (squares) + 1 DNA triangle, which should form a common structure.
In the first experiment 2 DNA tiles and 1 triangle DNA participated, the experiment itself was carried out at a constant temperature. Structurally, the triangle could have either the same binding domain as the squares, or it could have an additional binding domain complementary to the adjacent square.
In the first version of the experiment, 2 squares remain connected with each other, while the triangle is connected to one of the squares. In the second variant, the triangle replaced one of the squares, displacing it, so to speak. This has shown in practice that it is possible to change complex DNA origami. But now we need to make sure that this process is controlled by the scientist, and does not occur by itself at the expense of natural forces.
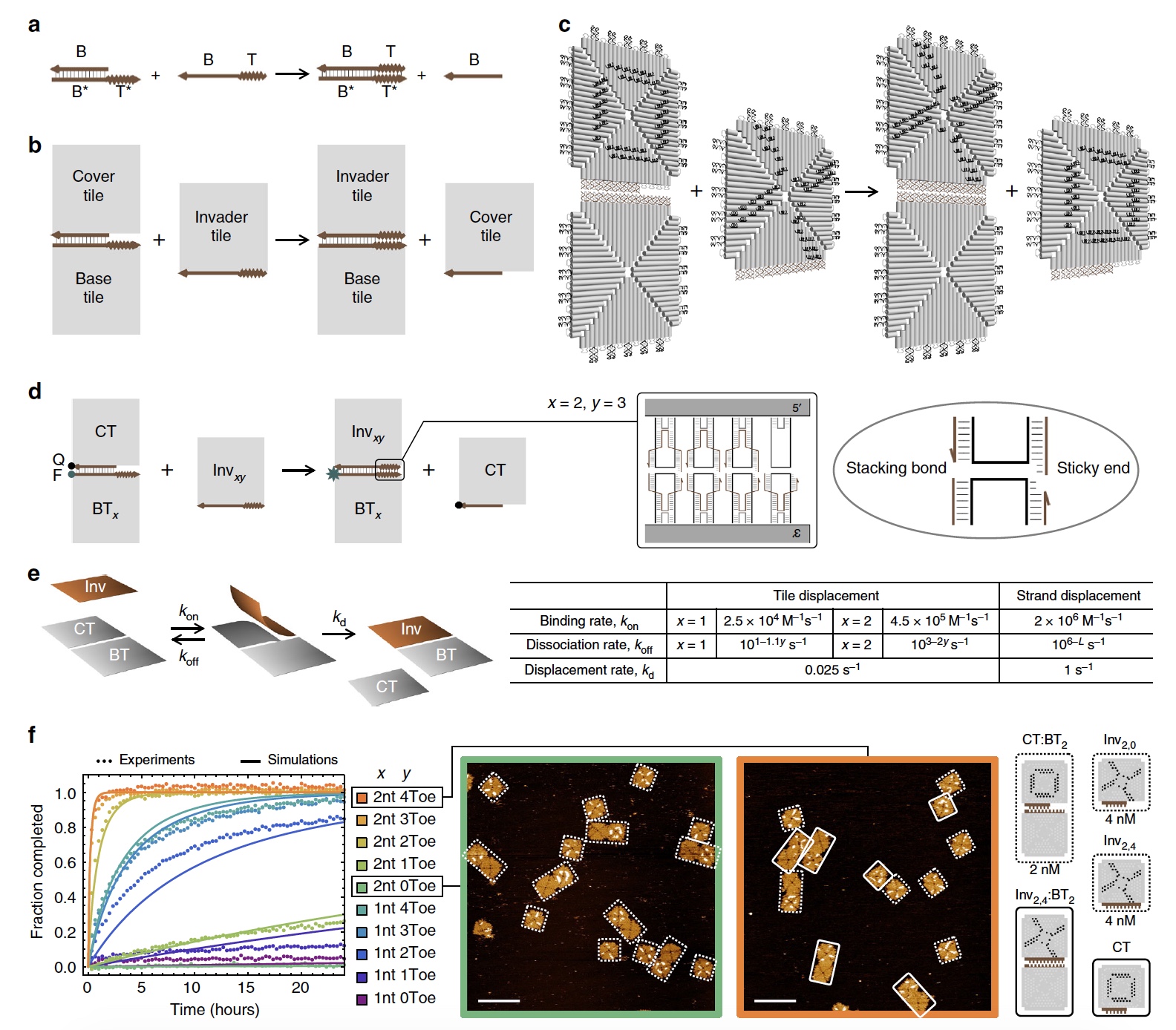
Image №1
If we talk about the displacement reactions of the DNA chains, then here we have a chain with a binding domain that connects to the "free" complementary domain. So we get a double-stranded structure in which the process of branch migration takes place when the chains move through the connection point ( 1a ). A similar process proceeds in the reactions of the DNA shift of the tiles (1b and 1c ).
From the 1c image, we can also see that each DNA tile consists of 4 isosceles triangles connected by braces, which we already know about.
Regarding the game of tic-tac-toe: for more convenient visualization of the process of displacement and reconfiguration, participants were marked with a cross (X) and a zero (O) by means of brackets of two different colors. Look at image No. 1, X (black) and O (white) are visible on the tiles.
Kinetics of Tile DNA Offset Reactions
In simple words, to achieve control over the kinetics of the process, it is necessary to make some reactions faster and others slower. In this way, the connection, disconnection and reconfiguration of the tiles can be achieved. But it is necessary to determine the range of kinetics, allowing it to carry out. For this, scientists have created connection points (between tiles) with different strength, so to speak, hitches.
The number of staples on the connecting branch migration in the domain was the same for all tiles. But the domain of the reference (connecting) points ranged from 0 to 4 ( 1d ).
The experiment involved the main tile (all 4 communication points are “active”) and accessory tiles (the number of communication points varied from 0 to 4). At the end of the domain of migration of branches, 2 pairs of brackets were modified by a fluorophore and a quencher. If the adnexal tile retains its connection with the main one, then the fluorophore will be extinguished, which will result in a low fluorescent signal. But if the accessory tile (with the quencher) is disconnected, the fluorescent signal will increase. Thus it will be possible to determine how the process proceeds.
A day later, it was possible to draw the first conclusions from the observations. Scientists note that the fluorescent trajectories of the accessory tiles with 0 connecting brackets remained almost unchanged. It was also noted that tiles with identical length of connecting edge, but with a large number of connecting points, exhibit much faster kinetics. It is curious that connection points with staples with 1-nucleotide and 2-nucleotide show a very different saturation (saturation) rate.
Image 1e shows the mathematical model used for the numerical evaluation of kinetics. Scientists themselves say that the model is very simple and is often used to evaluate the kinetics in the reactions of DNA strand bias, therefore it is also suitable for DNA tiles.
Comparative Analysis ( 1fa) simulations (simulated reactions) and experiments made it possible to determine the parameters that affect the recombination of DNA tiles.
First of all, scientists note that the speed of joining tiles is 10-100 times (depending on the length of the edge of joining a tile) less than the speed of joining DNA strands. A common feature between the displacement of tiles and filaments is that with an increase in the number of nucleotides at the points of connection, the decay rate exponentially decreases. At the same time, this rate is 40 times less for tiles than for DNA strands. The maximum total offset for DNA tiles (reaction rate) was 4.5x10 5 M -1 * s -1. If the concentration is low (<50 nM), the rate of bimolecular binding will limit the rate of displacement of the tiles. If the concentration is more than 50 nanomoles, then the rate of monomolecular displacement will limit the rate of displacement of the tiles.
Next, scientists conducted several experiments describing 3 main types of reconfiguration: competitive, consistent and cooperative. And now let's consider the results of experiments for each of them in more detail.
Competitive reconfiguration
To achieve competitive reconfiguration, scientists proposed using a sigmoidal function in response to signal concentration, thus obtaining a reconfiguration system based on information. This function is an important element of digital logical computation of DNA strand bias processes, therefore it can be used for DNA tiles too.
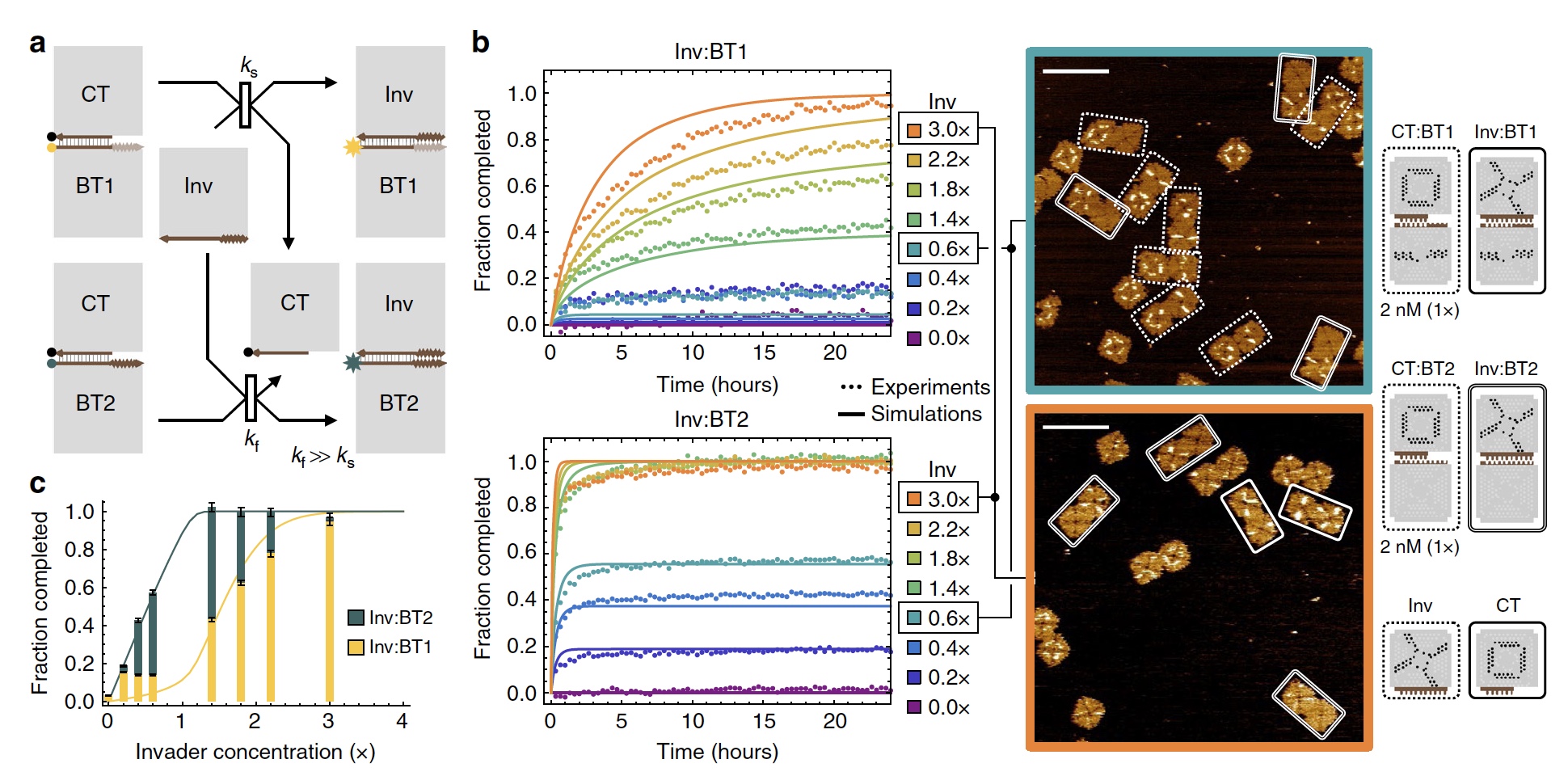
Image No. 2
In order to demonstrate the “work” of a sigmoidal function, scientists have created 2 competing tile displacement reactions that are activated by the same sub-tile. The difference in their speed ( 2a ).
In accessory tiles, the link point is four 2-nucleotide brackets. It is expected to connect to a suitable link point and with four 1-nucleotide straps. The reaction rate of the accessory tiles differs by 18 times. When the accessory tile had a concentration of less than 2 nM, a faster reaction was started ( 2b ). If the concentration was more than 2 nM, then a slower reaction started.
After 24 hours, it was found that the result of the faster reaction increased linearly, and the result of the slower reaction showed a sigmoidal function ( 2c ).
Sequential reconfiguration
Following competitive reconfiguration was consistent. This type is much more complicated, but gives more control over the process, unless of course everything works out.
So, scientists have created an array of 2x2, in which the first adnexal tile displaces another tile, freeing the previously used connection point. Further, the second adnexal structure of 2 tiles shifts 2 tiles from the array. A kind of cascade offset ( 3a ).
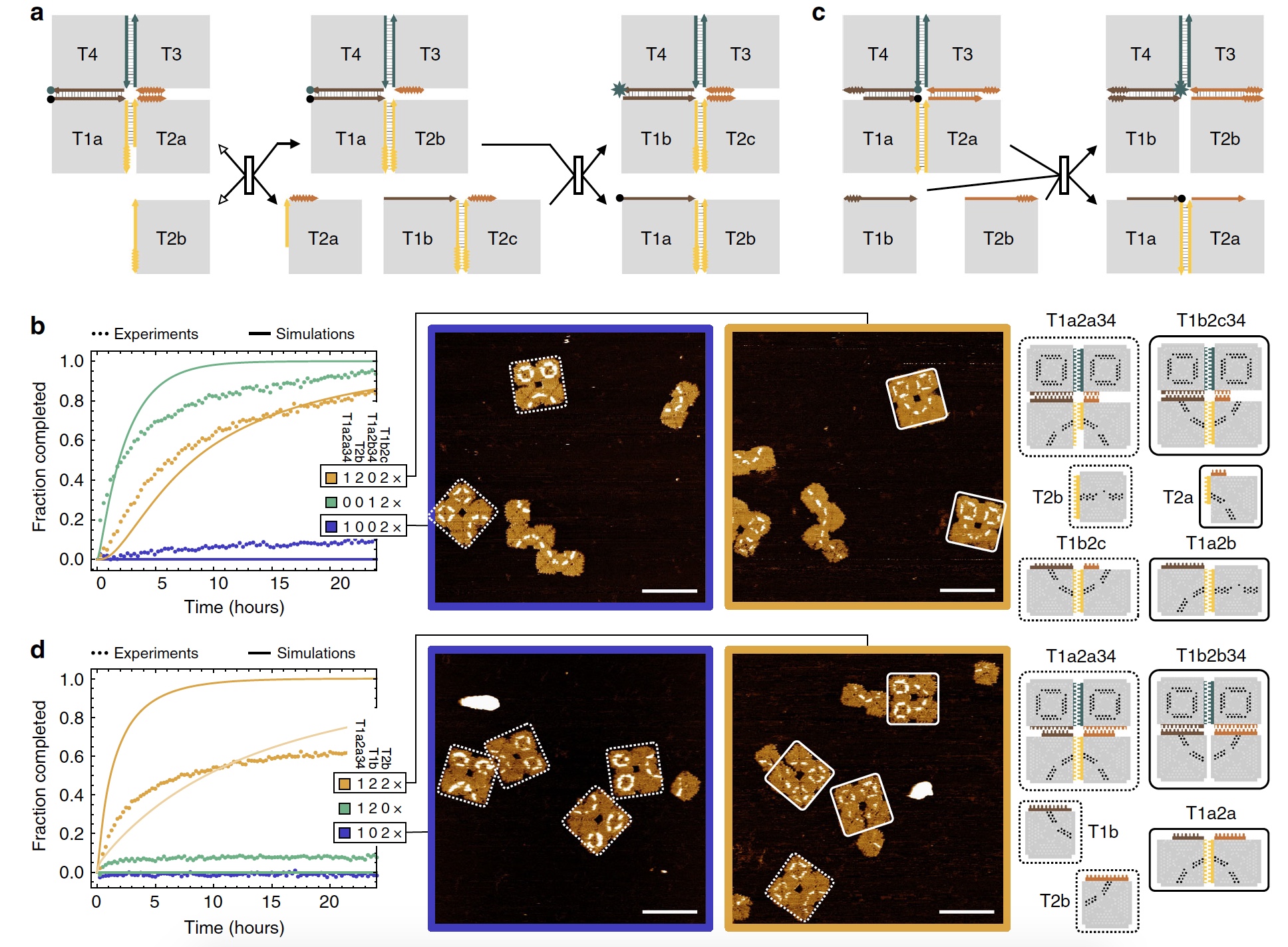
Image No. 3
Scientists have noticed that the displacement at the corners of the tiles is slow, about 100 times slower than in the rest of the edge of the tile. Therefore, such intermediate states of displacement must be taken into account for more complex reconfiguration procedures.
In image 3b we can see quite successful results of the experiment with sequential reconfiguration (orange curve).
And now a little more creativity from the researchers. See the images above (3b ): arrays that reacted to the first additional tile, but not to the second, in the end look like a “sad face”. But the arrays, in which the connection with both additional tiles was successful, look like a “smiling face”. And you say scientists have no sense of humor.
According to scientists, the more complex the structure of the reaction participants, the more difficult the process of reconfiguration itself, but the more interesting its result. The use of two types of participants (a 2x2 array and a double-tile structure) showed the following results on offset efficiency after 48 hours of observations: 83.3 ± 9.8% and 90.5 ± 6.1%. In other words, displacement reactions are excellent. There are some errors, but they are insignificant.
Experiments with sequential reconfiguration allowed not only to understand the basic principles of the sequence of communication points and the "cascade" of the reaction, but also demonstrated new features. The system is able to respond to more than 1 signal, indicating which instructions require execution and what is available for this. This signal is presented in the experiment in the form of two types of accessory structures. Even if the first subordinate structure comes into contact with the base before the second, the structure still shows the expected reconfiguration.
These processes for the most part proceed on their own. Scientists only slightly control them, but do not intervene in full. The next step in the experiments is to increase the control over the reconfiguration process and to study the possibility of programming this process.
Cooperative reconfiguration
In this experiment, there were 2 accessory tiles that were connected to two edges of a 2x2 ( 3c ) array . If there is only one tile, then it should move to the center of the array. In this case, the absence of a second tile allows you to reverse the process, thereby disconnecting the advent tile from the array. With both accessory tiles, two branches migrate at once, which converge in the center of the array, and as a result we get a cooperative shift.
Experience with fluorescence showed that if there is only one adventitious tile, the fluorescent signal is very weak, but it is enhanced if both tiles are present ( 3d : the blue and green curve is a weak signal, the yellow is a strong signal).
As in the previous experiment, the arrays, which interact only with 1 accessory tile, were marked as “sad face”, and the arrays and both tiles - “smiling face”. The latter were formed in 68.0 ± 7.7% of cases.
Tic-tac-toe
And, finally, the experiment "tic-tac-toe". In this experiment, a 3x3 array was used, which allows for 9 unique displacement of tiles in any order ( 4a ).
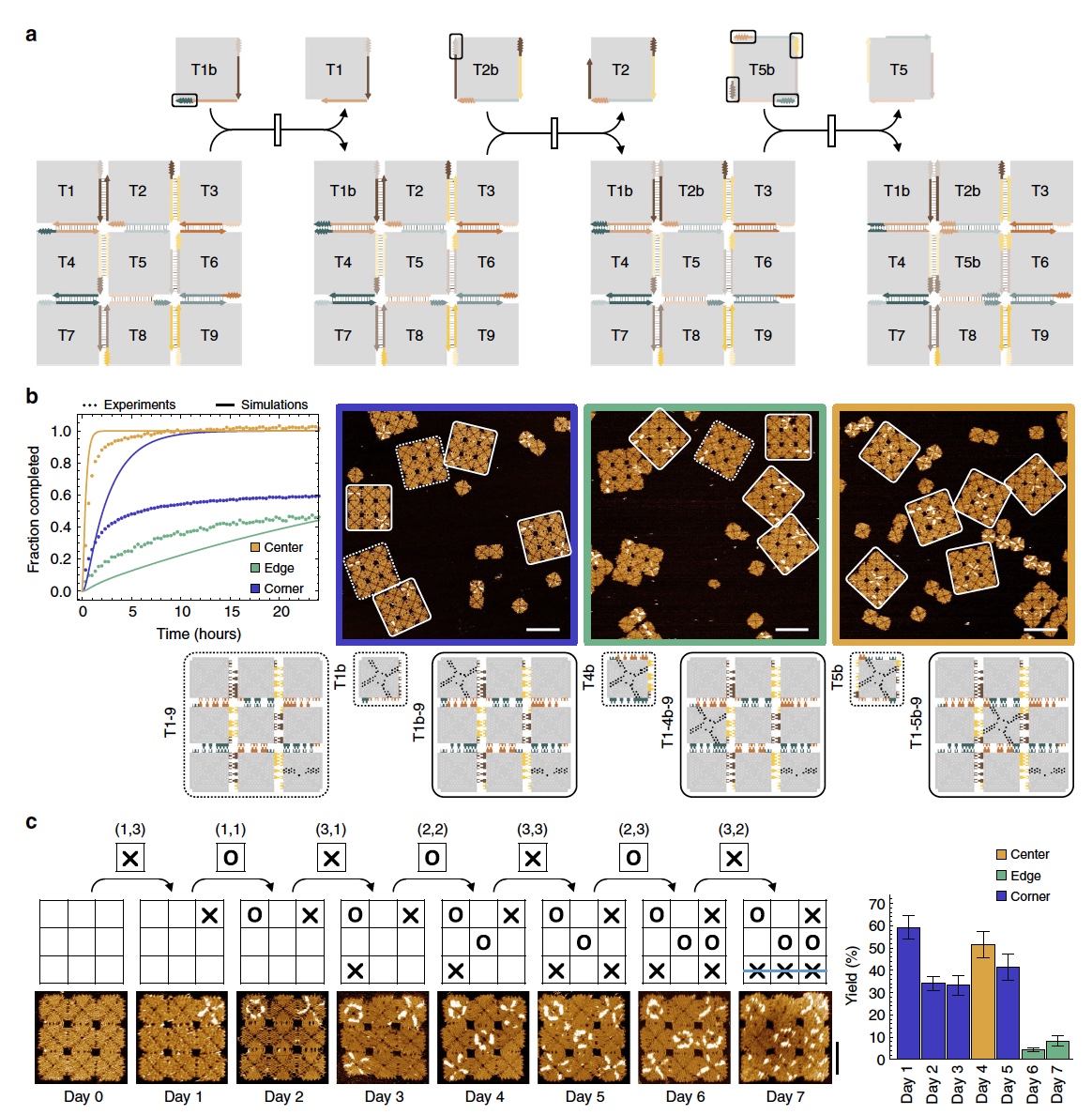
Image number 4
This experiment is a combination of several types of reactions at once: the offset of the corner tile, the offset of the edge tile and the offset of the central one. Along the perimeter of the array between each adjacent tiles there was one connection point, a total of 8 such points. 4 of them participated in the offset of the corner tile, when the accessory tile with the corresponding connection point is in contact with the edge tile near the corner tile. At this point, the branch migrates inside the tile edge and through a 90 ° angle, and the corner tile previously connected to the array is released and integrated into the array itself (inside the array when it was previously on the edge). The other 4 connection points were used to activate the offset of the edge tiles.
The most difficult thing in this experiment was to initiate the displacement of the central tile, due to the limited number of available communication points. Therefore, 4 additional points were added between the central tile and all adjacent ones ( 4a ).
Observations showed an excellent result. All accessory tiles were introduced into the array, which was rebuilt as planned by scientists. The conversion percentage inside the array was as follows: 78.4 ± 6.0% for the offset of the corner tiles, 52.8 ± 6.0% for the offset of the edge tiles, and 100% for the center tiles.
And now let's proceed directly to the game itself, which combines all types of reaction in the form of a cascade. The game board was an array of 3x3, all the tiles were “clean”, that is, not marked.
Each of the players, of which there were 2 naturally, got at their disposal 9 accessory tiles labeled "X" and "O". Making a move in this game is adding this tile to the test chamber at intervals of 24 hours (a long batch in tic-tac-toe, isn't it?).
Video demonstration of the game of tic-tac-toe using DNA tiles.
Observations ( 4b and 4c ) showed that the playing field reacted to all moves, that is, the reconfiguration proceeded in accordance with the instructions from the players (adding flagged tiles).
The whole problem was that by the end of the game there were a lot of accessory tiles on the field, respectively, the probability of their spontaneous displacement increased. Thus, the accuracy of the reconfiguration was also reduced. By the end of the game, this indicator was only 8.3 ± 2.3%. However, the result was still achieved, as we can see from the images above.
For a more detailed acquaintance with the nuances of research and experiments, I strongly recommend to look into the report of scientistsand additional materials to it.
Epilogue
The game of tic-tac-toe DNA tiles was carried out not for fun, but as a method of demonstrating very serious research that reveals the possibility of manipulating the displacement of elements in nanostructures built from DNA.
Of course, much remains to be polished: the number of accessory plates affecting the final reconfiguration rate, reaction rate, displacement accuracy, expansion of possible displacement options and, of course, an increase in the dimensions of the reconfigurable DNA structure.
Now many scientists are paying more and more attention to highly non-standard and non-trivial mechanisms for performing processes that have already become standard. Storage and processing of information, diagnosis and treatment of diseases, the study of complex systems, including biological ones - all these processes can be performed using DNA, since this incredibly small object hides in itself incredible properties and characteristics that we have yet to discover.
By the way, it turns out that if you enter the “tic-tac-toe” query into google, you can play this game. :) (I don’t know for whom how, but I didn’t notice it before)
And, of course, Friday offtop (I will continue the New Year theme, if you do not mind):
A few milots under the tree :)
Thank you for your attention, stay curious and have a great weekend, guys.
A few milots under the tree :)
Thank you for your attention, stay curious and have a great weekend, guys.
Thank you for staying with us. Do you like our articles? Want to see more interesting materials? Support us by placing an order or recommending to friends, 30% discount for Habr's users on a unique analogue of the entry-level servers that we invented for you: The whole truth about VPS (KVM) E5-2650 v4 (6 Cores) 10GB DDR4 240GB SSD 1Gbps from $ 20 or how to share the server? (Options are available with RAID1 and RAID10, up to 24 cores and up to 40GB DDR4).
VPS (KVM) E5-2650 v4 (6 Cores) 10GB DDR4 240GB SSD 1Gbps until spring for free if you pay for a period of six months, you can order here .
Dell R730xd 2 times cheaper? Only we have 2 x Intel Dodeca-Core Xeon E5-2650v4 128GB DDR4 6x480GB SSD 1Gbps 100 TV from $ 249in the Netherlands and the USA! Read about How to build an infrastructure building. class c using servers Dell R730xd E5-2650 v4 worth 9000 euros for a penny?
