How to create a 3D model of a human virus. Part one: collecting and analyzing scientific data

In the denouement of Blade Runner, the hero of Rutger Hauer says he saw a lot that people cannot even imagine - space battles, flaming warships - but is it really so difficult to imagine all this for a modern person spoiled by computer graphics? At the same time, we are surrounded by many things that we will never be able to consider in detail, due to fundamental physical limitations. Moreover, we ourselves are made up of such things. However, the good news is that these objects can be modeled and visualized using the currently developed 3D tools. And if you assemble a team in which there will be not only three-dimensional modelers, visualizers and designers, but also scientists, you can bring the result as close as possible to reality.
Under the cut, the first part of the story about our experience in creating scientifically valid virus models.
The world of molecular machines and viruses offers a ton of interesting challenges to CG teams. The problem is that so far there is no universal scientific technique that would allow to fully describe the structure of the viral particle. In order to describe the structure of the virus, you need to use many methods that give an idea of the individual pieces of the final puzzle. Electron microscopy allows you to evaluate the size and shape of virions, X-ray analysis can describe individual proteins or their fragments, and molecular biological and biochemical methods provide information on how many molecules are part of the virus and how they interact with each other. This creates a somewhat paradoxical situation: many viruses have been studied in great detail and in detail, but there are no images,
For example, modern electron micrographs of influenza virus particles look like this ( source ).
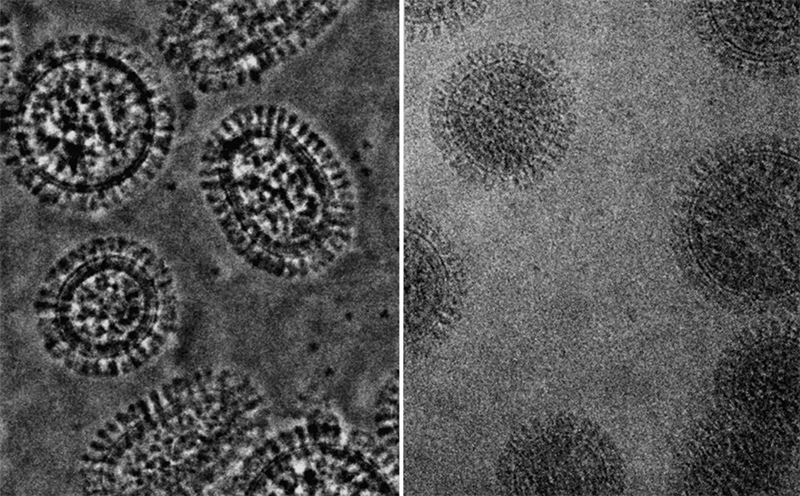
Data visualization cryoelectron microscopy complex genomic influenza virus A and reconstruction RNA packaging (yellow band) proteins B and C . The work with these data was published at the end of 2012 in the journal Science by a group of virologists from Madrid who helped us in creating a model of the A / H1N1 influenza virus.
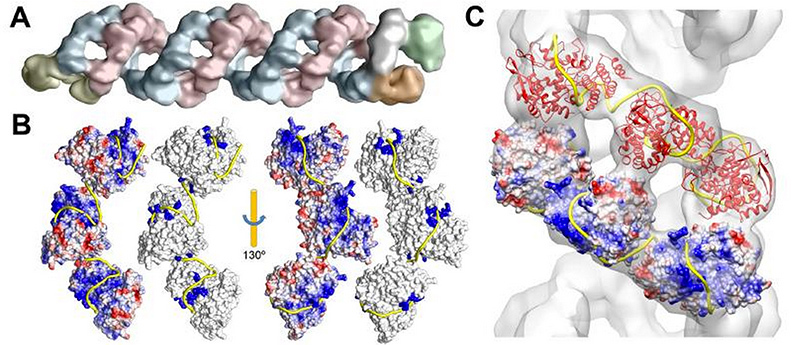
Collecting all available information is technically possible. But its systematization, processing and translation into a 3D model requires a team approach. At the same time, even a competent scientific consultant cannot possess the full baggage of highly specialized knowledge on the topic, therefore it is important to involve scientists who have devoted their entire career to working with a particular virus to the project. A modeler without biological education will not understand published scientific data and protein structures from Protein Data Bank, and also will not be able to correctly complete molecular models using molecular dynamics, where necessary (approximately 80-90% of the proteins that we encounter have an incomplete description of the spatial structure by 10-90%). A scientist, even having all the information separately, cannot assemble and visualize the complete model in professional programs for three-dimensional modeling. In our experience, only the close interaction of these specialists can give a neat and informative result.
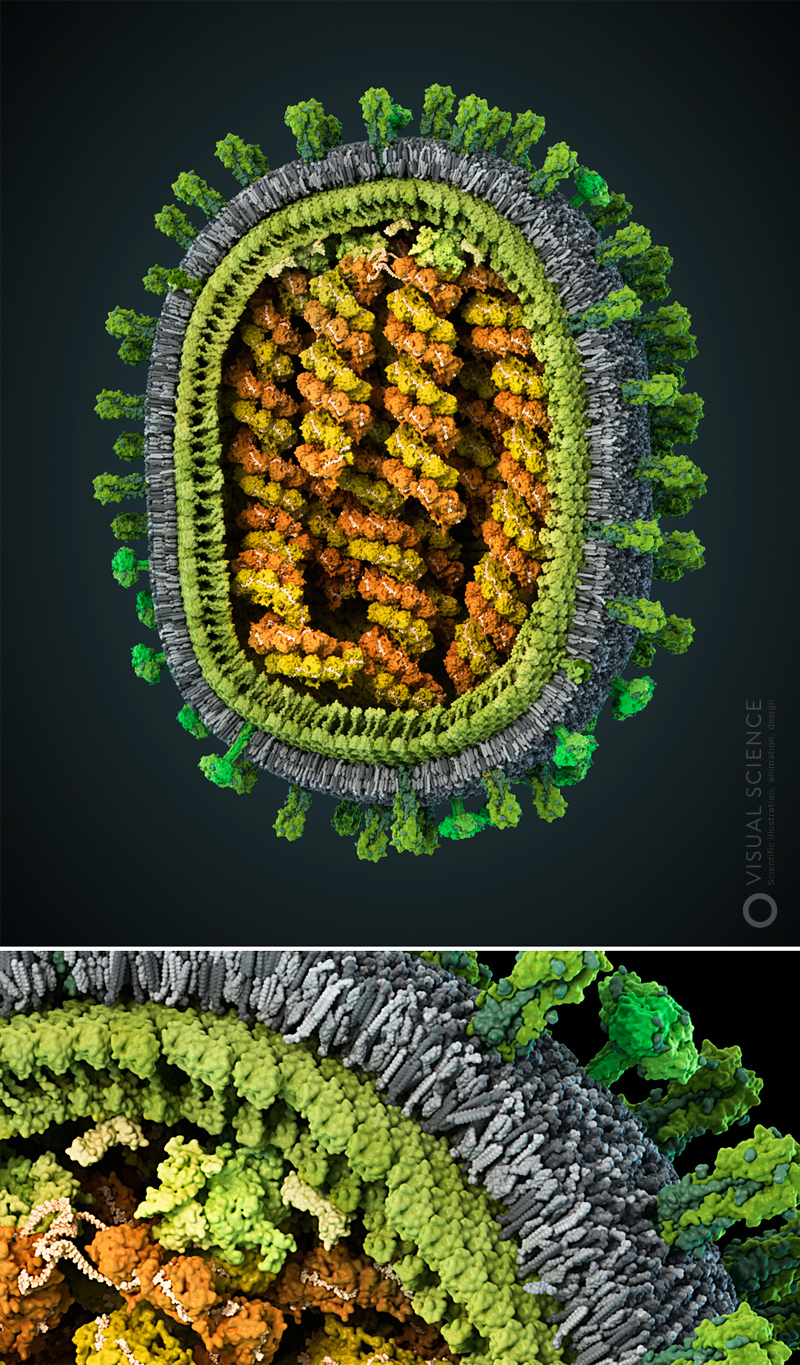
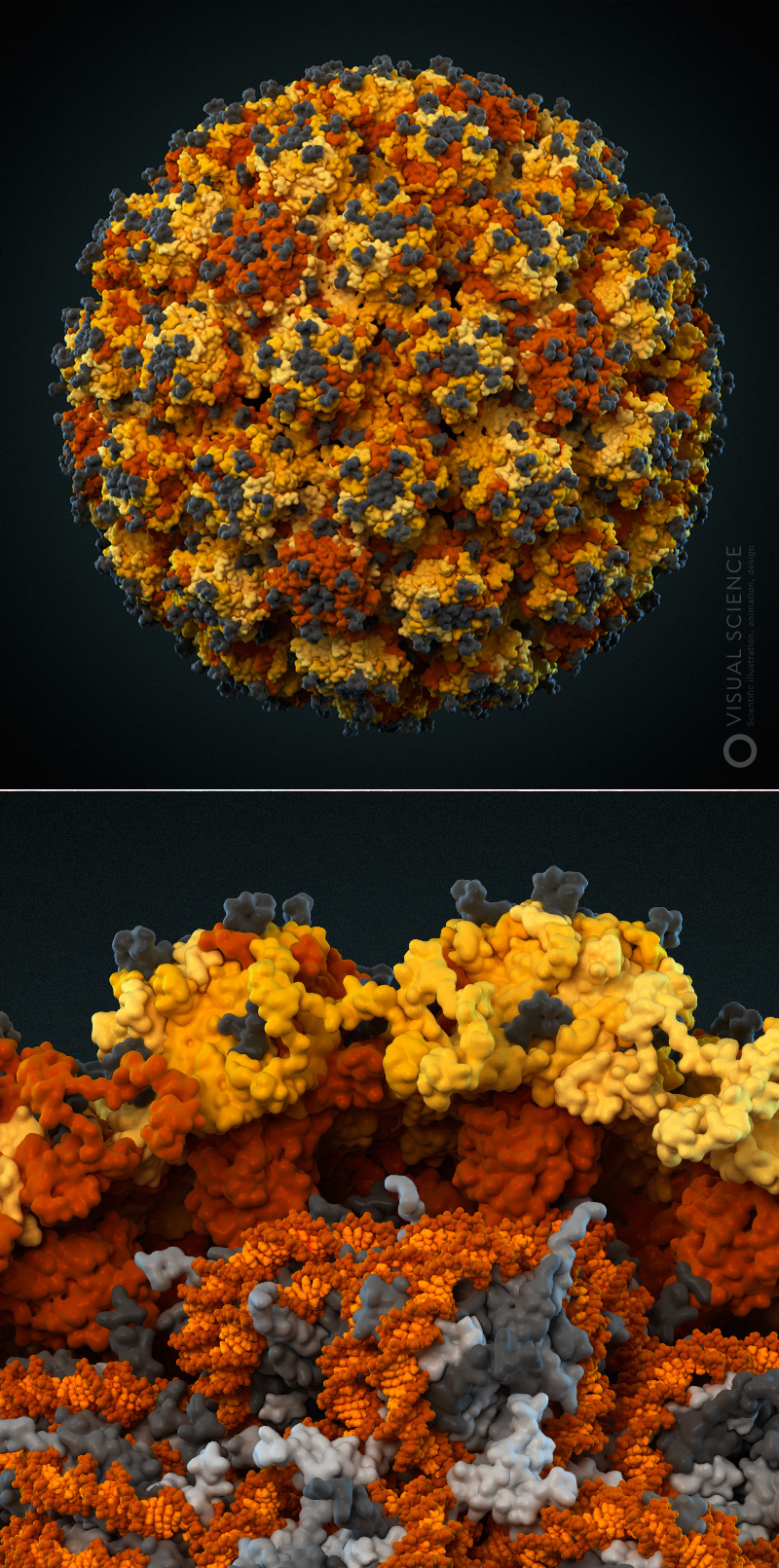
Image of influenza virus with detail to atoms. All proteins and protein complexes in the composition of the particle, as well as their quantitative ratios and position correspond to the data published in the scientific literature ( signatures of all components) The model was created with the participation of Jaime Martin-Benito and colleagues (Spanish National Center for Biotechnology, Madrid, Spain). year 2013.
The internal structure of the human immunodeficiency virus. The edge of the membrane membrane is visible, the proteins present inside the virion, capsid and fragments of the RNA of the virus enclosed in it ( signatures of all components ). The model was created with the participation of Egor Voronin (Global HIV Vaccine Enterprise). Prize for the best scientific illustration at the Science and Engineering Visualization Challenge in 2011.

A model of the proposed folding of the human papillomavirus genome. The model was created with the participation of Christopher Buck (National Cancer Institute, USA). year 2012.
Particle and individual proteins of the Ebola virus. The model was created with the participation of Ronald Harty (University of Pennislania, USA). Honorable mention the 2010 Science and Engineering Visualization Challenge. Exposition of the Salon of the Association of Medical Illustrators in Toronto in 2012.
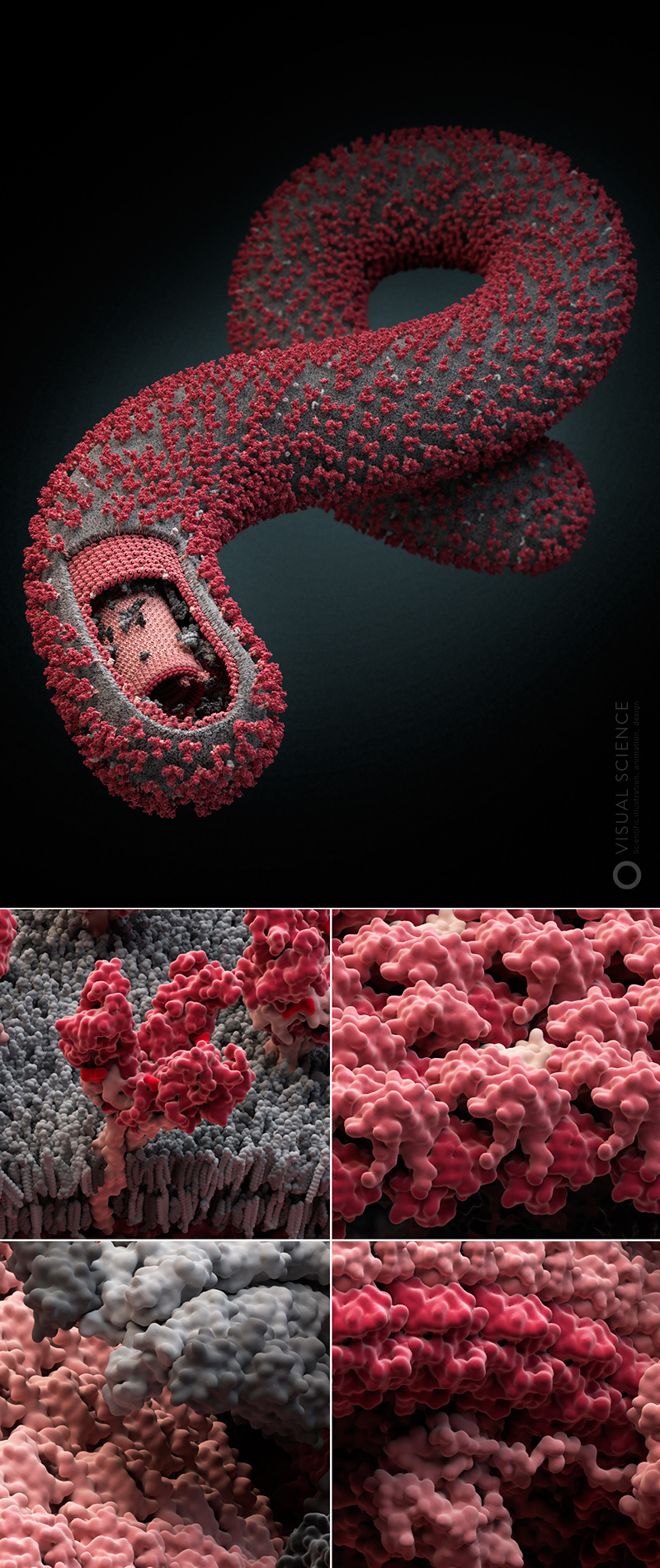
Our studio launched a non-profit project several years ago, the essence of which is to model and visualize the most common and dangerous human viruses. We named it Viral Park, or “Virus Zoo.” The project so far includes four viral models, several more are in development, and plans to make a series of about twenty virions. During the work on the project, we managed to master and adjust the process, highlighting a number of stages in it:
- Literature review and systematization of detected data
- Molecular Modeling and Dynamics
- Assembling a complete model from individual elements
- 3D visualization and design
- Creating materials based on the model from posters to applications, widgets and plastic models.
In this post we will talk a little about the first stage of our work.
Gathering information about the topic being studied is a task that scientists are constantly solving. It is impossible to make a new project without knowing what was published before you. For this, it is necessary to find and analyze first the review, and then the research publications on the issue of interest. The same scheme works when information about the structure of viruses is collected. Thanks to the databases of natural science publications of the main world magazines PubMed and Google Scholar, this process can be organized very efficiently. If you need background information about the biology of the virus, you can use the Viral Zone website and a lot of data on individual proteins is available in the Uniprot database. The structures of proteins or their fragments obtained by different teams of scientists using methods of nuclear magnetic resonance and X-ray diffraction analysis are available in the already mentioned Protein Data Bank in the form of coordinates of all atoms or, in some cases, only alpha atoms of a protein chain.
The task for the scientist in the process of creating a virus model is to collect, process and prepare all the information in a form that will be convenient for the rest of the team members. It is necessary to make a complete list of all types of molecules that form a particle, and all their interactions. In addition to proteins, these can be membrane lipids and viral genome molecules represented by DNA or RNA. Next, you need to understand in what quantities the molecules are present in the particle, and what places they occupy. This is the most difficult to search and often contradictory and incomplete information, since different methods can give different estimates. To clarify certain issues, we contact the authors of the articles in which they are discussed. This is an accepted practice in the scientific community, and scientists are often happy to
The result of a study of the literature should be the most detailed verbal picture of the future model. You need to understand what, in what quantities and how it is packaged in a viral particle. This can be reduced to a description, a table of quantities and interactions, and a plan of the model at the desired scale.
Further stages of work involve obtaining three-dimensional models of all the necessary components. One of the problems here is that not all proteins and their complexes can have atomic structures. Scientists simply have not yet been able to describe a significant portion of viral proteins. In our work, we use structural bioinformatics methods to fill this gap. We will talk about this in the following posts. We will also try to disclose details of how the complete model is assembled, its visualization and the creation of educational aids and widgets based on the result.
We believe that such a detailed approach to the modeling of molecular biological objects has great prospects from the point of view of its application in education, popularization of science and scientific communication. This model is also supported by the fact that such models get high marks at major international competitions of scientific illustration and design, positive reviews from famous colleagues, and it’s nice to include such images in their presentations even to Francoise Barre-Sinoussi , who received the Nobel Prize for discovering HIV.
In continuation of the topic, in addition to modeling viruses in the framework of the Virus Zoo, we will discuss the scope of scientific and medical illustration in general, talk about why this is relevant, how it differs from the growing scientific art, or Science Art, and how it will help make the world a better place. and science is clearer.
Only registered users can participate in the survey. Please come in.
Who is next?
- 14% Adenovirus 90
- 15.4% Hepatitis E 99
- 16.3% Stomatitis 105
- 11.8% Rhinovirus 76
- 40.4% Hepatitis C 259
- 17.9% Rotavirus 115
- 10.9% Coronavirus 70
- 43.9% Herpes 282
- 15.6% Rubella 100
- 18.5% Measles 119
