Deanonymization through genetic information
Brief summary:
Some time ago, on the Internet, databases on the genetic information of people (information of various levels of detail — from complete sequences ( sequence ) of the entire genome to limited information on short tandem repeats of the Y chromosome ( Y-STRs ) appeared on the Internet , for example, enthusiasts share information about their Y-STRs (haplotype) on genealogy sites to find out family ties and search for distant relatives, this data is not anonymous. Also anonymous medical genetic I have information, such as a research project " 1000 Genomes person"(a project to completely decipher the genomes of thousands of different people), where the anonymity of DNA donors is maintained for ethical reasons.
Here the fun begins. Genealogical databases (even very poorly populated, but nonetheless) make it possible to deanonymize people. For example, it is shown that in the case of artificial insemination with sperm from an anonymous donor, the use of genealogical databases allows you to find out at least the name of the real biological father of the child (that is, through very distant relatives who appeared in the database to find out which family the donor was from), and in the presence of additional information, such as place of residence, etc., allows you to uniquely identify the biological father. Recently it was shown that anonymous genetic data that is freely available, plus additional information about age, etc., can accurately establish the identities of approximately 50 anonymous DNA donors from the project “1000 Human Genomes”.

If you are interested in details and details, welcome to cat.
Some time ago, the theory was put forward that for accurate identification of a person in the modern world 33 bits of information are enough. By combining various information known about a person, we reduce information entropy, narrowing the circle of possible options, and, ultimately, accurately identifying a person. It is absolutely obvious that publicly available information about people - profiles of social networks, data provided during registration on numerous web services, etc., can directly help in deanonymizing a person registered in social networks. networks and other sites collecting user information. It is less obvious that data mining can reliably provide a lot of information about a person that is not directly indicated in his profile, based only on very indirect information, for example, on Facebook likes: www.pnas.org/content/110 / 15/5802. The authors of this work can correctly distinguish a Republican from a Democrat in 85% of cases, and a black American from a white in 95% of cases. However, there is another source of information about people that is not obvious to most IT people - open databases of genetic information. Their charm lies in the fact that the person does not have to leave his genetic information publicly available to be identified - his child’s genetic information is enough to identify through the very distant relatives (separated even 8 or more generations ago) that are lit up in the database at least the family from which the dad comes (and if there is additional information, find the dad himself). If your genetic information is publicly available, even if anonymously,
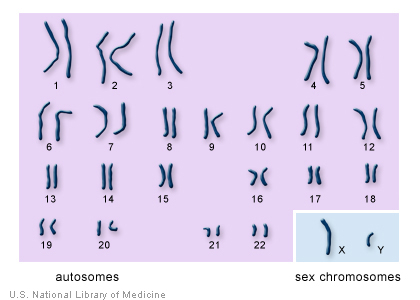
We all remember from school that in every cell of our body there is DNA in which our genetic information is encoded. In total, there are 46, or rather, 23 pairs of chromosomes in the cells of the human body, and each of our parents gets one 23 chromosome set. On the one hand, the genetic information of different people is quite similar: for example, if we take only the protein-coding regions of human DNA, then their similarity even to the protein-coding regions of chimpanzee DNA is approximately 99%, that is, they are even more similar among people . On the other hand, there are many areas in DNA that are not very genetically stable, for example, the so-called short tandem repeats ( STRs, short tandem repeats) These are sections of DNA in which a short sequence of 2-4 nucleotides in length is repeated more than 10 (up to a hundred) times. Such areas are not very convenient for copying, and cellular machinery for copying DNA often sculpts errors on these repeats. As a result, even the closest relatives have very similar, but nonetheless slightly different DNA sequences in these repeats. The farther the kinship is, the more differences accumulate, while the number of differences can even suggest how many generations ago there was a separation between these branches of the family. Another detail is interesting. Among 23 pairs of chromosomes there is a special pair - the so-called sex chromosomes, XX in women and XY in men. Here inheritance works as follows - since a woman has the XX genotype, the female egg always carries the X chromosome. The sperm can carry either the X or Y chromosome, since in men the genotype is XY. Thus, the sex of the child is always determined by the sperm and there is another interesting property - the Y chromosome is always inherited along the male line, from grandfather to father, from father to son, etc. (The X chromosome of the same man comes from the mother and can be inherited as from a paternal grandmother, and from a maternal grandmother, statistically with a probability of 50%). Since the surname usually comes to us from the father, short tandem repeats located on the Y-chromosome ( from father to son, etc. (the X-chromosome of the same man comes from mom and can be inherited from both grandmother by father and grandmother by mother, statistically with a 50% probability). Since the surname usually comes to us from the father, short tandem repeats located on the Y-chromosome ( from father to son, etc. (the X-chromosome of the same man comes from mom and can be inherited from both grandmother by father and grandmother by mother, statistically with a 50% probability). Since the surname usually comes to us from the father, short tandem repeats located on the Y-chromosome (en.wikipedia.org/wiki/Y-STR ), inherited only from father to son, can be used to establish a person’s surname. Although in principle STRs from any chromosomes can be used to assess the degree of kinship, the Y chromosome, due to the nature of its inheritance, allows one to clearly outline male inheritance.
Now let's talk about what level of detail is available genetic information. Of course, the most complete genetic information is the complete sequence ( sequence ) of the entire genome . The first Human Genome project lasted 13 years, with a project budget of approximately $ 3 billion (!). However, in the course of this work, technologies were developed and brought to mind that later made it possible to sequence the genome of one person in 2 months and a million dollars. Although in recent years the cost of sequencing has continued to plummet and has not yet reached the bottom (the task is to reach the cost of $ 1,000 per gene), nevertheless, it is still an expensive pleasure, inaccessible to ordinary people. But the project " 1000 human genomes" was launched . Its goal is to compile a fairly complete catalog of genetic differences between people, for this anonymous DNA samples were taken from people of different races, from different countries, in order to get the greatest variety of genomes within this thousand. At the moment, the main part of sequencing is completed and anonymous genomic sequences are freely available: www.1000genomes.org/homeso that scientists can easily analyze and compare them (that's where the endless work for data mining is!). In full sequence, even now, there is much to be said about a person - to determine his predisposition to many diseases, such as cancer or Alzheimer's disease, to reliably determine his race, in many cases to reliably determine eye color, and so on - a correlation has recently been found between some genetic markers and level of education .
Hybridization methods, for example, on- chip hybridization, are less informative, but much cheaper and faster.. In this case, we do not determine the complete sequence of the entire genome, but ask which of the known possible variants is present in this particular genome? There is such a scary word SNP - single nucleotide polymorphismOne of the goals of the “1000 Human Genomes” project is precisely to search for and characterize such single nucleotide polymorphisms (when the DNA sequence of two people differs in one place by one nucleotide). Many of the SNPs are genetic markers of predisposition to cancer and other diseases, therefore, for medical purposes it is not necessary to sequence the entire genome - it is enough to drive hybridization on a chip, see which SNPs for which it is known that they are markers of certain diseases are present in the patient and make conclusion about his predisposition to these diseases. SNP databases are also publicly available .
Even less informative, but sufficient to identify a person, is information about his haplotype - the features of his short tandem repeats (STRs), which we have already discussed above. Technically, the STRs profile can be obtained very cheaply in less than half a day using PCR , or you can again use the hybridization method on a chip . Although the haplotype is unlikely to help with medical diagnosis, it allows you to identify a person that has long been used in forensics, and all advanced countries - the United States , Britain and, including, our homelandhave already taken care of compiling genetic databases for identifying criminals. These databases, as I understand it, are closed and not accessible to the general public. It should be noted that, based on more complete information, it is possible to restore a less complete one, i.e., according to the full genomic sequence, STR profiles can be easily counted with the corresponding tools , a similar operation is theoretically possible for data on polymorphism (SNP).
We pass to the most interesting. As we have already said, the inheritance of the Y chromosome goes strictly from father to son, which means that the Y-STR data ( haplotype ) allows you to track the paternal line of inheritance. At present, at least 8 genealogical databases are available on the Web, containing a total of hundreds of thousands of records comparing the haplotype with the person’s last name. The largest open access and searchable databases are Ysearch ( www.ysearch.org ) and SMGF ( www.smgf.org) The idea of these databases is to search for your distant relatives and dig in your own family tree - for this you make your haplotype available for search in the hope that distant relatives will someday be found, plus you yourself look for similar (related) among the haplotypes already available in the database. To determine the haplotype, you need to send your DNA sample for analysis to one of these companies. It should be noted that basically these bases cover the population of the USA and Western countries, therefore everything that will be said below about identification of the person will be true more likely for the West. Say, in Ysearch there were as many as 11 people with the surname “Ivanov”, one from Bulgaria, two Russians, the rest with “double” origin Russia-USA, or the origin is not indicated. People with the surname "Johns" (probably
What can be done with these databases now? With a low occupancy - hundreds of thousands of records, the available data already makes it possible to determine membership in a certain family for millions (maybe tens of millions) of people, which already makes up a significant percentage for the population of the same USA (population ~ 300 million). The method is sensitive enough to draw relatives from family lines that split more than 8 generations ago, and the bases are constantly growing, increasing coverage. Now the era of genomic anonymity (for example, in the case of anonymous sperm donation) is coming to an end. More and more sentimental stories like this. In this case, a woman made artificial insemination in a medical facility using sperm from an anonymous donor and gave birth to a wonderful girl with some mental disorders. Although the institution signed a non-disclosure obligation of the donor and strictly abided by this agreement, it did not save the donor from deanonymization. First of all, the mother, using genetic databases, was able to find several other children born from the same anonymous donor - there is even a special base aimed at reuniting brothers and sisters on the father of an anonymous sperm donor - www.donorsiblingregistry.com. It turned out that many of the children (and there were already 13 of them) have autism and other abnormalities. Our heroine was very interested in it and she persuaded another woman who gave birth to a boy from the same donor father to take her son's genetic material for analysis through the Y-STRs database. As a result, two families of very distant relatives of the donor were found from the databases, but with the help of additional information that the donor discovered about himself (he allowed women to give a minimum of information about themselves - their education, mother’s profession and that his father was a famous baseball player) he was uniquely identified - He had to receive guests and get acquainted with his daughter and her mom.
Another story- if in short, on his deathbed, the old man told his son that in fact he was not his own, but adopted. The old man died, and the son had an idea-fix to find his real father. The details are similar to the previous case - the Y-STRs base, a clue through a distant relative, promotion with additional information about the place of residence, etc., as a result, the biological father was found, though it turned out that he had already passed away. But the paternal brothers are alive and now a happy person regularly visits them to listen to stories about his biological daddy. And there will be more and more such stories as the databases are filled.
There is another side to this coin. As we discussed above, the full genetic sequences of anonymous DNA donors, for example, from the project “1000 Human Genomes,” are now publicly available on the Web. In addition to the actual complete sequence of genomes, some minimum information about donors is available - age, geographic location, etc. The personalities of these people should remain secret for ethical reasons - they donated DNA in the name of science, so that the genetic differences between people can be analyzed as accurately as possible, but their DNA also contains sensitive information - their predisposition to diseases, for example. I would not want insurance companies to know this information (in America you can’t get sick with insurance medicine). However, according to the complete sequence of the genome, you can safely count Y-STRs, and then ... Well, you understand. Base,Science about deanonymization of at least 50 out of 1,000 people. Ethical committees sigh heavily, guys from the 1000 Human Genomes project urgently remove age information from all records to make accurate identification impossible (the search circle is narrowed to a few (tens) people , but more precisely, it won’t work out anymore), and the rest recall the story of a simple African-American Henrietta Lax .
Her story is sad and somewhat surreal. She died of cervical cancer at the age of 31 in 1951, having lived an outstanding life (perhaps the most outstanding thing she did was to give birth to a child at 14), however today every molecular biologist knows at least the first two letters of her name and surnames - HeLa. This is the name of the most famous cancer cell line used today for experiments in most laboratories working in the field of cancer research and not only. The point is that when the doctor took Henrietta Lax's cancer cell for analysis, he noticed that they quickly shared and were relatively easy to cultivate. Many cancer cell lines are unstable, the same line turned out to be very stable, for which she was loved. Henrietta herself died more than 60 years ago, but the cancer that killed her is still alive in hundreds of laboratories. Since cancer cells are human cells of their own, in which the control of cell divisions has broken down, it can be said that Henrietta herself, in a sense, is still alive in the form of her cancer. Now she is immortal, lives in laboratories and multiplies and multiplies in a nutrient medium ... The story, worthy of Stephen King. Of course, when sequencing technologies were brought to the modern level, the complete genomic sequence of the HeLa cancer line was determined and, of course, made publicly available. The European Laboratory of Molecular Biology (EMBL, Germany), which carried out the project, said that the publication of the complete genomic sequence of the HeLa line does not reveal any information about Henrietta Lax herself and her relatives and descendants, but we all know that all markers of disease susceptibility are fully sequencing - in full view, and American insurers are already peeking over their shoulders. After numerous The European Laboratory of Molecular Biology (EMBL, Germany), which carried out the project, said that the publication of the complete genomic sequence of the HeLa line does not reveal any information about Henrietta Lax herself and her relatives and descendants, but we all know that all markers of disease susceptibility are fully sequencing - in full view, and American insurers are already peeking over their shoulders. After numerous The European Laboratory of Molecular Biology (EMBL, Germany), which carried out the project, said that the publication of the complete genomic sequence of the HeLa line does not reveal any information about Henrietta Lax herself and her relatives and descendants, but we all know that all markers of disease susceptibility are fully sequencing - in full view, and American insurers are already peeking over their shoulders. After numerousraids EMBL has recognized that the publication of the genome sequence violates the privacy Henriette relatives and remove it from the free access.
To summarize. Databases appeared on the Web for searching for distant relatives and genealogy games for genetic information (alas, to fully use the search, you will have to send them a sample of your genetic information for analysis - usually this is scraping from the inside of the cheek, from which they will then extract the DNA). These databases can be effectively used not only to search for lost cousins, but also to accurately identify a sperm donor in case of artificial insemination (anonymous donor, forget about anonymity) or biological parents in case of adoption. Now the databases contain hundreds of thousands of records, which will allow tens of millions of people to be pulled through distant kinship (if additional information is available for accurate identification). This figure will only grow with an increase in the number of records in the databases. The question of deanonymization through anonymous genetic information used in biological and medical research is also raised. So, if you inadvertently left someone your genetic information in the form of a child - they may come to you and say: “Hello, dad!”, And if you participated in anonymous medical genetic research - do not be surprised if the insurers become breaking crazy money for your health insurance, since you have a marker of predisposition to skin cancer or something like that.
On this optimistic note, allow me to take my leave - be healthy, do not get sick, and be careful about your genetic information.
Some time ago, on the Internet, databases on the genetic information of people (information of various levels of detail — from complete sequences ( sequence ) of the entire genome to limited information on short tandem repeats of the Y chromosome ( Y-STRs ) appeared on the Internet , for example, enthusiasts share information about their Y-STRs (haplotype) on genealogy sites to find out family ties and search for distant relatives, this data is not anonymous. Also anonymous medical genetic I have information, such as a research project " 1000 Genomes person"(a project to completely decipher the genomes of thousands of different people), where the anonymity of DNA donors is maintained for ethical reasons.
Here the fun begins. Genealogical databases (even very poorly populated, but nonetheless) make it possible to deanonymize people. For example, it is shown that in the case of artificial insemination with sperm from an anonymous donor, the use of genealogical databases allows you to find out at least the name of the real biological father of the child (that is, through very distant relatives who appeared in the database to find out which family the donor was from), and in the presence of additional information, such as place of residence, etc., allows you to uniquely identify the biological father. Recently it was shown that anonymous genetic data that is freely available, plus additional information about age, etc., can accurately establish the identities of approximately 50 anonymous DNA donors from the project “1000 Human Genomes”.

If you are interested in details and details, welcome to cat.
Introduction
Some time ago, the theory was put forward that for accurate identification of a person in the modern world 33 bits of information are enough. By combining various information known about a person, we reduce information entropy, narrowing the circle of possible options, and, ultimately, accurately identifying a person. It is absolutely obvious that publicly available information about people - profiles of social networks, data provided during registration on numerous web services, etc., can directly help in deanonymizing a person registered in social networks. networks and other sites collecting user information. It is less obvious that data mining can reliably provide a lot of information about a person that is not directly indicated in his profile, based only on very indirect information, for example, on Facebook likes: www.pnas.org/content/110 / 15/5802. The authors of this work can correctly distinguish a Republican from a Democrat in 85% of cases, and a black American from a white in 95% of cases. However, there is another source of information about people that is not obvious to most IT people - open databases of genetic information. Their charm lies in the fact that the person does not have to leave his genetic information publicly available to be identified - his child’s genetic information is enough to identify through the very distant relatives (separated even 8 or more generations ago) that are lit up in the database at least the family from which the dad comes (and if there is additional information, find the dad himself). If your genetic information is publicly available, even if anonymously,
A bit of genetics

We all remember from school that in every cell of our body there is DNA in which our genetic information is encoded. In total, there are 46, or rather, 23 pairs of chromosomes in the cells of the human body, and each of our parents gets one 23 chromosome set. On the one hand, the genetic information of different people is quite similar: for example, if we take only the protein-coding regions of human DNA, then their similarity even to the protein-coding regions of chimpanzee DNA is approximately 99%, that is, they are even more similar among people . On the other hand, there are many areas in DNA that are not very genetically stable, for example, the so-called short tandem repeats ( STRs, short tandem repeats) These are sections of DNA in which a short sequence of 2-4 nucleotides in length is repeated more than 10 (up to a hundred) times. Such areas are not very convenient for copying, and cellular machinery for copying DNA often sculpts errors on these repeats. As a result, even the closest relatives have very similar, but nonetheless slightly different DNA sequences in these repeats. The farther the kinship is, the more differences accumulate, while the number of differences can even suggest how many generations ago there was a separation between these branches of the family. Another detail is interesting. Among 23 pairs of chromosomes there is a special pair - the so-called sex chromosomes, XX in women and XY in men. Here inheritance works as follows - since a woman has the XX genotype, the female egg always carries the X chromosome. The sperm can carry either the X or Y chromosome, since in men the genotype is XY. Thus, the sex of the child is always determined by the sperm and there is another interesting property - the Y chromosome is always inherited along the male line, from grandfather to father, from father to son, etc. (The X chromosome of the same man comes from the mother and can be inherited as from a paternal grandmother, and from a maternal grandmother, statistically with a probability of 50%). Since the surname usually comes to us from the father, short tandem repeats located on the Y-chromosome ( from father to son, etc. (the X-chromosome of the same man comes from mom and can be inherited from both grandmother by father and grandmother by mother, statistically with a 50% probability). Since the surname usually comes to us from the father, short tandem repeats located on the Y-chromosome ( from father to son, etc. (the X-chromosome of the same man comes from mom and can be inherited from both grandmother by father and grandmother by mother, statistically with a 50% probability). Since the surname usually comes to us from the father, short tandem repeats located on the Y-chromosome (en.wikipedia.org/wiki/Y-STR ), inherited only from father to son, can be used to establish a person’s surname. Although in principle STRs from any chromosomes can be used to assess the degree of kinship, the Y chromosome, due to the nature of its inheritance, allows one to clearly outline male inheritance.
Technical Details: Different levels of detail on genetic information
Now let's talk about what level of detail is available genetic information. Of course, the most complete genetic information is the complete sequence ( sequence ) of the entire genome . The first Human Genome project lasted 13 years, with a project budget of approximately $ 3 billion (!). However, in the course of this work, technologies were developed and brought to mind that later made it possible to sequence the genome of one person in 2 months and a million dollars. Although in recent years the cost of sequencing has continued to plummet and has not yet reached the bottom (the task is to reach the cost of $ 1,000 per gene), nevertheless, it is still an expensive pleasure, inaccessible to ordinary people. But the project " 1000 human genomes" was launched . Its goal is to compile a fairly complete catalog of genetic differences between people, for this anonymous DNA samples were taken from people of different races, from different countries, in order to get the greatest variety of genomes within this thousand. At the moment, the main part of sequencing is completed and anonymous genomic sequences are freely available: www.1000genomes.org/homeso that scientists can easily analyze and compare them (that's where the endless work for data mining is!). In full sequence, even now, there is much to be said about a person - to determine his predisposition to many diseases, such as cancer or Alzheimer's disease, to reliably determine his race, in many cases to reliably determine eye color, and so on - a correlation has recently been found between some genetic markers and level of education .
Hybridization methods, for example, on- chip hybridization, are less informative, but much cheaper and faster.. In this case, we do not determine the complete sequence of the entire genome, but ask which of the known possible variants is present in this particular genome? There is such a scary word SNP - single nucleotide polymorphismOne of the goals of the “1000 Human Genomes” project is precisely to search for and characterize such single nucleotide polymorphisms (when the DNA sequence of two people differs in one place by one nucleotide). Many of the SNPs are genetic markers of predisposition to cancer and other diseases, therefore, for medical purposes it is not necessary to sequence the entire genome - it is enough to drive hybridization on a chip, see which SNPs for which it is known that they are markers of certain diseases are present in the patient and make conclusion about his predisposition to these diseases. SNP databases are also publicly available .
Even less informative, but sufficient to identify a person, is information about his haplotype - the features of his short tandem repeats (STRs), which we have already discussed above. Technically, the STRs profile can be obtained very cheaply in less than half a day using PCR , or you can again use the hybridization method on a chip . Although the haplotype is unlikely to help with medical diagnosis, it allows you to identify a person that has long been used in forensics, and all advanced countries - the United States , Britain and, including, our homelandhave already taken care of compiling genetic databases for identifying criminals. These databases, as I understand it, are closed and not accessible to the general public. It should be noted that, based on more complete information, it is possible to restore a less complete one, i.e., according to the full genomic sequence, STR profiles can be easily counted with the corresponding tools , a similar operation is theoretically possible for data on polymorphism (SNP).
Luke, who is your father?
We pass to the most interesting. As we have already said, the inheritance of the Y chromosome goes strictly from father to son, which means that the Y-STR data ( haplotype ) allows you to track the paternal line of inheritance. At present, at least 8 genealogical databases are available on the Web, containing a total of hundreds of thousands of records comparing the haplotype with the person’s last name. The largest open access and searchable databases are Ysearch ( www.ysearch.org ) and SMGF ( www.smgf.org) The idea of these databases is to search for your distant relatives and dig in your own family tree - for this you make your haplotype available for search in the hope that distant relatives will someday be found, plus you yourself look for similar (related) among the haplotypes already available in the database. To determine the haplotype, you need to send your DNA sample for analysis to one of these companies. It should be noted that basically these bases cover the population of the USA and Western countries, therefore everything that will be said below about identification of the person will be true more likely for the West. Say, in Ysearch there were as many as 11 people with the surname “Ivanov”, one from Bulgaria, two Russians, the rest with “double” origin Russia-USA, or the origin is not indicated. People with the surname "Johns" (probably
What can be done with these databases now? With a low occupancy - hundreds of thousands of records, the available data already makes it possible to determine membership in a certain family for millions (maybe tens of millions) of people, which already makes up a significant percentage for the population of the same USA (population ~ 300 million). The method is sensitive enough to draw relatives from family lines that split more than 8 generations ago, and the bases are constantly growing, increasing coverage. Now the era of genomic anonymity (for example, in the case of anonymous sperm donation) is coming to an end. More and more sentimental stories like this. In this case, a woman made artificial insemination in a medical facility using sperm from an anonymous donor and gave birth to a wonderful girl with some mental disorders. Although the institution signed a non-disclosure obligation of the donor and strictly abided by this agreement, it did not save the donor from deanonymization. First of all, the mother, using genetic databases, was able to find several other children born from the same anonymous donor - there is even a special base aimed at reuniting brothers and sisters on the father of an anonymous sperm donor - www.donorsiblingregistry.com. It turned out that many of the children (and there were already 13 of them) have autism and other abnormalities. Our heroine was very interested in it and she persuaded another woman who gave birth to a boy from the same donor father to take her son's genetic material for analysis through the Y-STRs database. As a result, two families of very distant relatives of the donor were found from the databases, but with the help of additional information that the donor discovered about himself (he allowed women to give a minimum of information about themselves - their education, mother’s profession and that his father was a famous baseball player) he was uniquely identified - He had to receive guests and get acquainted with his daughter and her mom.
Another story- if in short, on his deathbed, the old man told his son that in fact he was not his own, but adopted. The old man died, and the son had an idea-fix to find his real father. The details are similar to the previous case - the Y-STRs base, a clue through a distant relative, promotion with additional information about the place of residence, etc., as a result, the biological father was found, though it turned out that he had already passed away. But the paternal brothers are alive and now a happy person regularly visits them to listen to stories about his biological daddy. And there will be more and more such stories as the databases are filled.
Using anonymous genetic information
There is another side to this coin. As we discussed above, the full genetic sequences of anonymous DNA donors, for example, from the project “1000 Human Genomes,” are now publicly available on the Web. In addition to the actual complete sequence of genomes, some minimum information about donors is available - age, geographic location, etc. The personalities of these people should remain secret for ethical reasons - they donated DNA in the name of science, so that the genetic differences between people can be analyzed as accurately as possible, but their DNA also contains sensitive information - their predisposition to diseases, for example. I would not want insurance companies to know this information (in America you can’t get sick with insurance medicine). However, according to the complete sequence of the genome, you can safely count Y-STRs, and then ... Well, you understand. Base,Science about deanonymization of at least 50 out of 1,000 people. Ethical committees sigh heavily, guys from the 1000 Human Genomes project urgently remove age information from all records to make accurate identification impossible (the search circle is narrowed to a few (tens) people , but more precisely, it won’t work out anymore), and the rest recall the story of a simple African-American Henrietta Lax .
Her story is sad and somewhat surreal. She died of cervical cancer at the age of 31 in 1951, having lived an outstanding life (perhaps the most outstanding thing she did was to give birth to a child at 14), however today every molecular biologist knows at least the first two letters of her name and surnames - HeLa. This is the name of the most famous cancer cell line used today for experiments in most laboratories working in the field of cancer research and not only. The point is that when the doctor took Henrietta Lax's cancer cell for analysis, he noticed that they quickly shared and were relatively easy to cultivate. Many cancer cell lines are unstable, the same line turned out to be very stable, for which she was loved. Henrietta herself died more than 60 years ago, but the cancer that killed her is still alive in hundreds of laboratories. Since cancer cells are human cells of their own, in which the control of cell divisions has broken down, it can be said that Henrietta herself, in a sense, is still alive in the form of her cancer. Now she is immortal, lives in laboratories and multiplies and multiplies in a nutrient medium ... The story, worthy of Stephen King. Of course, when sequencing technologies were brought to the modern level, the complete genomic sequence of the HeLa cancer line was determined and, of course, made publicly available. The European Laboratory of Molecular Biology (EMBL, Germany), which carried out the project, said that the publication of the complete genomic sequence of the HeLa line does not reveal any information about Henrietta Lax herself and her relatives and descendants, but we all know that all markers of disease susceptibility are fully sequencing - in full view, and American insurers are already peeking over their shoulders. After numerous The European Laboratory of Molecular Biology (EMBL, Germany), which carried out the project, said that the publication of the complete genomic sequence of the HeLa line does not reveal any information about Henrietta Lax herself and her relatives and descendants, but we all know that all markers of disease susceptibility are fully sequencing - in full view, and American insurers are already peeking over their shoulders. After numerous The European Laboratory of Molecular Biology (EMBL, Germany), which carried out the project, said that the publication of the complete genomic sequence of the HeLa line does not reveal any information about Henrietta Lax herself and her relatives and descendants, but we all know that all markers of disease susceptibility are fully sequencing - in full view, and American insurers are already peeking over their shoulders. After numerousraids EMBL has recognized that the publication of the genome sequence violates the privacy Henriette relatives and remove it from the free access.
Finita la commedia
To summarize. Databases appeared on the Web for searching for distant relatives and genealogy games for genetic information (alas, to fully use the search, you will have to send them a sample of your genetic information for analysis - usually this is scraping from the inside of the cheek, from which they will then extract the DNA). These databases can be effectively used not only to search for lost cousins, but also to accurately identify a sperm donor in case of artificial insemination (anonymous donor, forget about anonymity) or biological parents in case of adoption. Now the databases contain hundreds of thousands of records, which will allow tens of millions of people to be pulled through distant kinship (if additional information is available for accurate identification). This figure will only grow with an increase in the number of records in the databases. The question of deanonymization through anonymous genetic information used in biological and medical research is also raised. So, if you inadvertently left someone your genetic information in the form of a child - they may come to you and say: “Hello, dad!”, And if you participated in anonymous medical genetic research - do not be surprised if the insurers become breaking crazy money for your health insurance, since you have a marker of predisposition to skin cancer or something like that.
On this optimistic note, allow me to take my leave - be healthy, do not get sick, and be careful about your genetic information.
