Part number 5. Folding bio-calculations. One fundamental problem
In this article, we will look at how to fold a single helix in RNA. For understanding, you need to read all the previous parts From proteins to RNA , Mat. Criteria , How to reduce the number of turns of the circuit? , How to evaluate the progress of folding single- RNA? , Limitation of optimizing methods in games with and without adversary . If earlier everything went smoothly for us, then here we will face a serious problem. Can someone tell me how to solve it.
Let us repeat the drawing of the secondary structure of the ribozyme that we will be folding.
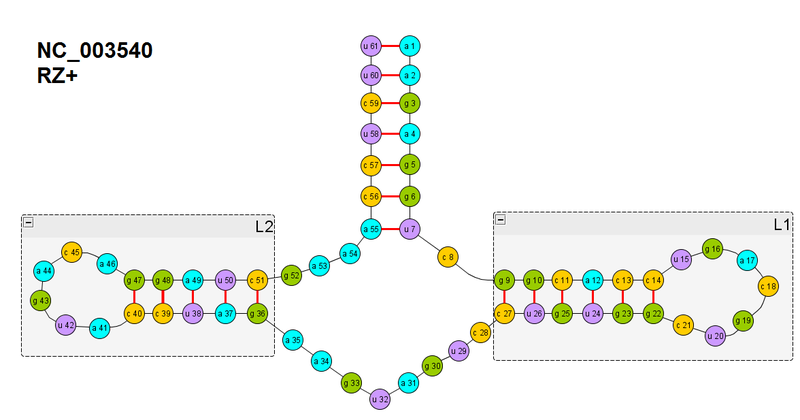
Here we pay attention to the spiral L1. In the previous part, we introduced the objective function for the formation of hydrogen bonds between one pair of nucleotides (let's call it F (xy), where xy is the pair of nucleotides). Here we need to form as many as six: between nucleotides 14-22, 13-23, 12-24, 11-25, 10-26, 9-27. When we form them, the RNA chain coils into a spiral by itself -> this is the geometric arrangement of the atoms that form the hydrogen bonds.
It would seem that we can take and use such an objective function:
Fo = F (14-22) + F (13-23) + F (12-24) + F (11-25) + F (10-26) + F ( 9-27)
But it turns out that the bonds at the end of the spiral, i.e. between nucleotides 10-26, 9-27 will form faster. Why? Well, in general, it’s clear that the chain length is longer for the ends, there are more opportunities to dock, the probability of finding the necessary turns faster is greater.
But how does it interfere? The fact is, as soon as a hydrogen bond is formed, the atoms come so close that it then becomes so difficult for them to find a turn so that on the contrary they move away from each other - there is a high probability of collision. And this is not physics - it is geometry in those restrictions about which I wrote earlier.
So, if bonds 10-26, 9-27 were almost formed, then under the condition “do not break these bonds”, and our objective function assumes this, it almost nullifies the possibility of forming bonds at the beginning of the chain, i.e. between nucleotides 14-22, 13-23.
Of course, this is due to calculations, and not to nature. Everything is good in nature.
But how the chain rotates. In the FoldIt game - there is such an opportunity - the Rebuild tool (watch the video, it will become clearer what it is about). He fixes the end of the chain, and begins to rotate the nucleotides. Moreover, if we say we rotate 18 nucleotides, then all nucleotides up to it 1-17 remain motionless. But after turning 18 nucleotides will surely change their position all the rest further, i.e. 19-61. Moreover, the further they are from the 18th, the stronger they will change their location.
So, if we formed a link 9-27, and we want to form a link 14-22, then this is affected by the turns inside the loop 15, 16, 17, 18, 19, 20, 21 ... let's say the rotation X in the 18 nucleotide greatly reduces the target function F (14-22), but at the same time it breaks the connection 9-27, and the objective function F (9-27) increases very much. It is clear that with the general objective function Fo we will never carry out such a turn, which means we will not be able to move towards the intended goal.
In the game FoldIt there is such an opportunity to freeze nucleotides. And then the Rebuild tool rotates the chain of only non-frozen nucleotides (this is observed in the video above). For example, it can freeze nucleotides 9-27, i.e. where we formed a connection, and rotate inside between points 9 - 27 without moving them. How he does it? Please tell me.
I got out of this problem by changing the objective function Fo. It is visible on the slide.

The idea is very simple - to control the compression ratio which pairs of hydrogen bonds need to be formed at the beginning and which later. With coefficient = 1, this function turns into just the sum as in Fo.
This greatly improves the folding of the spiral. But there can still be loops, especially at short distances. Then you have to change the compression ratio, one way or another to break the connection and repeat the process. This is already being done manually, and it is impossible to automate.
upd. An important addition see below in the comments with 3 drawings of a folding ribozyme.
Let us repeat the drawing of the secondary structure of the ribozyme that we will be folding.

Here we pay attention to the spiral L1. In the previous part, we introduced the objective function for the formation of hydrogen bonds between one pair of nucleotides (let's call it F (xy), where xy is the pair of nucleotides). Here we need to form as many as six: between nucleotides 14-22, 13-23, 12-24, 11-25, 10-26, 9-27. When we form them, the RNA chain coils into a spiral by itself -> this is the geometric arrangement of the atoms that form the hydrogen bonds.
It would seem that we can take and use such an objective function:
Fo = F (14-22) + F (13-23) + F (12-24) + F (11-25) + F (10-26) + F ( 9-27)
But it turns out that the bonds at the end of the spiral, i.e. between nucleotides 10-26, 9-27 will form faster. Why? Well, in general, it’s clear that the chain length is longer for the ends, there are more opportunities to dock, the probability of finding the necessary turns faster is greater.
But how does it interfere? The fact is, as soon as a hydrogen bond is formed, the atoms come so close that it then becomes so difficult for them to find a turn so that on the contrary they move away from each other - there is a high probability of collision. And this is not physics - it is geometry in those restrictions about which I wrote earlier.
So, if bonds 10-26, 9-27 were almost formed, then under the condition “do not break these bonds”, and our objective function assumes this, it almost nullifies the possibility of forming bonds at the beginning of the chain, i.e. between nucleotides 14-22, 13-23.
Of course, this is due to calculations, and not to nature. Everything is good in nature.
But how the chain rotates. In the FoldIt game - there is such an opportunity - the Rebuild tool (watch the video, it will become clearer what it is about). He fixes the end of the chain, and begins to rotate the nucleotides. Moreover, if we say we rotate 18 nucleotides, then all nucleotides up to it 1-17 remain motionless. But after turning 18 nucleotides will surely change their position all the rest further, i.e. 19-61. Moreover, the further they are from the 18th, the stronger they will change their location.
So, if we formed a link 9-27, and we want to form a link 14-22, then this is affected by the turns inside the loop 15, 16, 17, 18, 19, 20, 21 ... let's say the rotation X in the 18 nucleotide greatly reduces the target function F (14-22), but at the same time it breaks the connection 9-27, and the objective function F (9-27) increases very much. It is clear that with the general objective function Fo we will never carry out such a turn, which means we will not be able to move towards the intended goal.
In the game FoldIt there is such an opportunity to freeze nucleotides. And then the Rebuild tool rotates the chain of only non-frozen nucleotides (this is observed in the video above). For example, it can freeze nucleotides 9-27, i.e. where we formed a connection, and rotate inside between points 9 - 27 without moving them. How he does it? Please tell me.
I got out of this problem by changing the objective function Fo. It is visible on the slide.

The idea is very simple - to control the compression ratio which pairs of hydrogen bonds need to be formed at the beginning and which later. With coefficient = 1, this function turns into just the sum as in Fo.
This greatly improves the folding of the spiral. But there can still be loops, especially at short distances. Then you have to change the compression ratio, one way or another to break the connection and repeat the process. This is already being done manually, and it is impossible to automate.
upd. An important addition see below in the comments with 3 drawings of a folding ribozyme.
