Varieties of SIMD
- Transfer
 During the development of meshoptimizer , the question often arises: “Can this algorithm be used with SIMD?” The
During the development of meshoptimizer , the question often arises: “Can this algorithm be used with SIMD?” The library is performance-oriented, but SIMD does not always provide significant speed advantages. Unfortunately, SIMD can make the code less portable and less maintainable. Therefore, in each case, it is necessary to seek a compromise. When performance is paramount, you have to develop and maintain separate SIMD implementations for the SSE and NEON instruction sets. In other cases, you need to understand what the effect of using SIMD is. Today we’ll try to speed up the sloppy mesh simplifier — a new algorithm recently added to the library — using the SSEn / AVXn instruction sets.
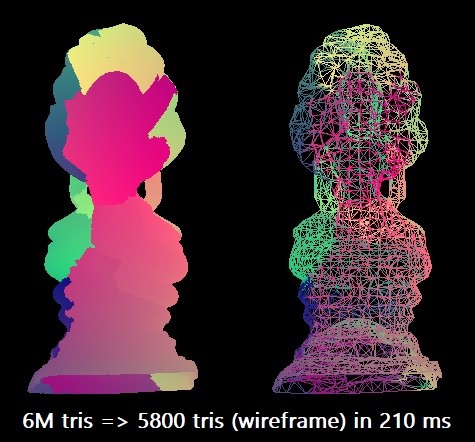
For our benchmark, we simplify the Thai Buddha model from 6 million triangles to 0.1% of this number. We use the Microsoft Visual Studio 2019 compiler for the target x64 architecture. The scalar algorithm can perform such rationalization in approximately 210 ms in a single Intel Core i7-8700K thread (at ~ 4.4 GHz). This corresponds to approximately 28.5 million triangles per second. Perhaps this is enough in practice, but I was curious to explore the maximum capabilities of the equipment.
In some cases, the procedure can be parallelized by dividing the grid into pieces, but for this it is necessary to conduct an additional analysis of connectivity in order to preserve the boundaries, so for now we will restrict ourselves to purely SIMD optimizations.
Seven Dimensions
To understand the possibilities for optimization, we will perform profiling using Intel VTune. Run the procedure 100 times to ensure that there is enough data.
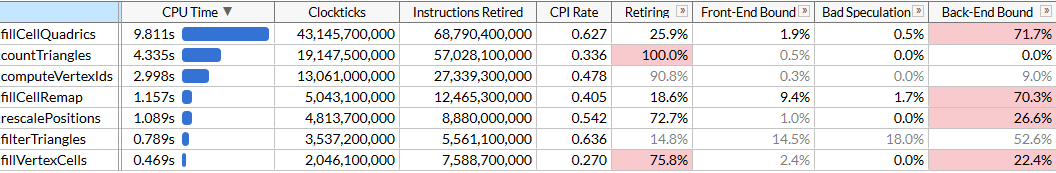
Here I turned on the microarchitecture mode to fix the execution time of each function, as well as to find bottlenecks. We see that rationalization is performed using a set of functions, each of which requires a certain number of cycles. The list of functions is sorted by time. Here they are in order of execution to make the algorithm easier to understand:
rescalePositionsnormalizes the positions of all vertices into a single cube in order to prepare for quantization usingcomputeVertexIdscomputeVertexIdscomputes a 30-bit quantized identifier for each vertex on a uniform grid of a given size, where each axis is quantized on a grid (grid size 10 bits, so the identifier is 30)countTrianglescalculates the approximate number of triangles that the optimizer will create for a given grid size, assuming the union of all the vertices in one grid cellfillVertexCellsfills in a table that correlates all the vertices with the corresponding cells; all vertices with the same ID correspond to one cellfillCellQuadricsfills the structureQuadric(4 × 4 symmetric matrix) for each cell to reflect aggregate information about the corresponding geometryfillCellRemapcalculates the vertex index for each cell, choosing one of the vertices in this cell, and minimizes geometric distortionfilterTrianglesdisplays the final set of triangles in accordance with the vertex-cell-vertex tables constructed earlier; naive translation can produce an average of ~ 5% duplicate triangles, so the function filters duplicates.
computeVertexIdsand they countTrianglesare executed several times: the algorithm determines the mesh size for merging vertices, performing an accelerated binary search to achieve the target number of triangles (in our case 6000) and calculates the number of triangles that each mesh size will generate at each iteration. Other functions are launched once. In our file, five search passes give a target mesh size of 40 3 . VTune obligingly reports that the most resource-intensive function is the one that calculates quadrics: it takes almost half the total execution time of 21 s. This is the first goal for optimizing SIMD.
SIMD piece by piece
Let's look at the source code
fillCellQuadricsto better understand what exactly it calculates:staticvoidfillCellQuadrics(Quadric* cell_quadrics, constunsignedint* indices, size_t index_count, const Vector3* vertex_positions, constunsignedint* vertex_cells){
for (size_t i = 0; i < index_count; i += 3)
{
unsignedint i0 = indices[i + 0];
unsignedint i1 = indices[i + 1];
unsignedint i2 = indices[i + 2];
unsignedint c0 = vertex_cells[i0];
unsignedint c1 = vertex_cells[i1];
unsignedint c2 = vertex_cells[i2];
bool single_cell = (c0 == c1) & (c0 == c2);
float weight = single_cell ? 3.f : 1.f;
Quadric Q;
quadricFromTriangle(Q,
vertex_positions[i0],
vertex_positions[i1],
vertex_positions[i2],
weight);
if (single_cell)
{
quadricAdd(cell_quadrics[c0], Q);
}
else
{
quadricAdd(cell_quadrics[c0], Q);
quadricAdd(cell_quadrics[c1], Q);
quadricAdd(cell_quadrics[c2], Q);
}
}
}The function iterates over all the triangles, calculates a quadric for each of them, and adds it to the quadrics of each cell. Quadric - a 4 × 4 symmetric matrix, represented as 10 floating-point numbers:
structQuadric
{float a00;
float a10, a11;
float a20, a21, a22;
float b0, b1, b2, c;
};Calculation of a quadric requires solving the plane equation for a triangle, constructing a quadratic matrix and weighing it using the area of the triangle:
staticvoidquadricFromPlane(Quadric& Q, float a, float b, float c, float d){
Q.a00 = a * a;
Q.a10 = b * a;
Q.a11 = b * b;
Q.a20 = c * a;
Q.a21 = c * b;
Q.a22 = c * c;
Q.b0 = d * a;
Q.b1 = d * b;
Q.b2 = d * c;
Q.c = d * d;
}
staticvoidquadricFromTriangle(Quadric& Q, const Vector3& p0, const Vector3& p1, const Vector3& p2, float weight){
Vector3 p10 = {p1.x - p0.x, p1.y - p0.y, p1.z - p0.z};
Vector3 p20 = {p2.x - p0.x, p2.y - p0.y, p2.z - p0.z};
Vector3 normal =
{
p10.y * p20.z - p10.z * p20.y,
p10.z * p20.x - p10.x * p20.z,
p10.x * p20.y - p10.y * p20.x
};
float area = normalize(normal);
float distance = normal.x*p0.x + normal.y*p0.y + normal.z*p0.z;
quadricFromPlane(Q, normal.x, normal.y, normal.z, -distance);
quadricMul(Q, area * weight);
}It looks like there are a lot of floating point operations, so they can be parallelized using SIMD. First, we represent each vector as a 4-wide SIMD vector, and also change the structure
Quadricto 12 floating point numbers instead of 10 so that it fits exactly into three SIMD registers (increasing the size does not affect performance) and change the order of the fields so that the calculations in quadricFromPlanebecome more uniform:structQuadric
{float a00, a11, a22;
float pad0;
float a10, a21, a20;
float pad1;
float b0, b1, b2, c;
};Here, some calculations, in particular the scalar product, are not very consistent with earlier versions of SSE. Fortunately, an instruction for scalar product appeared in SSE4.1, which is very useful for us.
staticvoidfillCellQuadrics(Quadric* cell_quadrics, constunsignedint* indices, size_t index_count, const Vector3* vertex_positions, constunsignedint* vertex_cells){
constint yzx = _MM_SHUFFLE(3, 0, 2, 1);
constint zxy = _MM_SHUFFLE(3, 1, 0, 2);
constint dp_xyz = 0x7f;
for (size_t i = 0; i < index_count; i += 3)
{
unsignedint i0 = indices[i + 0];
unsignedint i1 = indices[i + 1];
unsignedint i2 = indices[i + 2];
unsignedint c0 = vertex_cells[i0];
unsignedint c1 = vertex_cells[i1];
unsignedint c2 = vertex_cells[i2];
bool single_cell = (c0 == c1) & (c0 == c2);
__m128 p0 = _mm_loadu_ps(&vertex_positions[i0].x);
__m128 p1 = _mm_loadu_ps(&vertex_positions[i1].x);
__m128 p2 = _mm_loadu_ps(&vertex_positions[i2].x);
__m128 p10 = _mm_sub_ps(p1, p0);
__m128 p20 = _mm_sub_ps(p2, p0);
__m128 normal = _mm_sub_ps(
_mm_mul_ps(
_mm_shuffle_ps(p10, p10, yzx),
_mm_shuffle_ps(p20, p20, zxy)),
_mm_mul_ps(
_mm_shuffle_ps(p10, p10, zxy),
_mm_shuffle_ps(p20, p20, yzx)));
__m128 areasq = _mm_dp_ps(normal, normal, dp_xyz); // SSE4.1
__m128 area = _mm_sqrt_ps(areasq);
// masks the result of the division when area==0// scalar version does this in normalize()
normal = _mm_and_ps(
_mm_div_ps(normal, area),
_mm_cmpneq_ps(area, _mm_setzero_ps()));
__m128 distance = _mm_dp_ps(normal, p0, dp_xyz); // SSE4.1
__m128 negdistance = _mm_sub_ps(_mm_setzero_ps(), distance);
__m128 normalnegdist = _mm_blend_ps(normal, negdistance, 8);
__m128 Qx = _mm_mul_ps(normal, normal);
__m128 Qy = _mm_mul_ps(
_mm_shuffle_ps(normal, normal, _MM_SHUFFLE(3, 2, 2, 1)),
_mm_shuffle_ps(normal, normal, _MM_SHUFFLE(3, 0, 1, 0)));
__m128 Qz = _mm_mul_ps(negdistance, normalnegdist);
if (single_cell)
{
area = _mm_mul_ps(area, _mm_set1_ps(3.f));
Qx = _mm_mul_ps(Qx, area);
Qy = _mm_mul_ps(Qy, area);
Qz = _mm_mul_ps(Qz, area);
Quadric& q0 = cell_quadrics[c0];
__m128 q0x = _mm_loadu_ps(&q0.a00);
__m128 q0y = _mm_loadu_ps(&q0.a10);
__m128 q0z = _mm_loadu_ps(&q0.b0);
_mm_storeu_ps(&q0.a00, _mm_add_ps(q0x, Qx));
_mm_storeu_ps(&q0.a10, _mm_add_ps(q0y, Qy));
_mm_storeu_ps(&q0.b0, _mm_add_ps(q0z, Qz));
}
else
{
// omitted for brevity, repeats the if() body// three times for c0/c1/c2
}
}
}There is nothing particularly interesting in this code; we are abundantly using unaligned load / store instructions. Although Vector3's input can be aligned, there seems to be no noticeable penalty for unaligned reads. Please note that in the first half of the function vectors are not used, which is good - our vectors have three components, and in some cases only one (see areasq / area / distance calculation), while the processor performs 4 operations in parallel. In any case, let's see how parallelization has helped.

A hundred launches
fillCellQuadricsnow runs in 5.3 s instead of 9.8 s, which saves about 45 ms on each operation - not bad, but not very impressive. In many instructions, we use three instead of four components, as well as precise multiplication, which gives a fairly significant delay. If you previously wrote instructions for SIMD, then you know how to do the scalar product correctly. To do this, you need to do four vectors at once. Instead of storing one full vector in one SIMD register, we use three registers - in one we store four components
x, in the other it will be stored у, and in the third z. Here four vectors are needed for work at once: it means that we will process four triangles simultaneously.We have many arrays with dynamic indexing. Usually it helps to pre-transfer data to prepared component arrays
x/ y/ z(or rather, small SIMD registers are usually used, for example float x[8], y[8], z[8], for each of the 8 vertices in the input data: this is called AoSoA (arrays of array structures) and gives a good balance between cache locality and simplicity of loading into SIMD registers), but here dynamic indexing will not work very well, therefore, we will load the data for four triangles as usual, and transpose the vectors using a convenient macro _MM_TRANSPOSE. Theoretically, you need to calculate each component of four finite quadrics in its own SIMD register (for example, we will
__m128 Q_a00have four componentsa00finite quadrics). In this case, the operations on the quadrics fit quite well in the 4-wide SIMD instructions, and the conversion actually slows down the code - therefore, we transpose only the initial vectors, and then transpose the plane equations back and run the same code that we used to calculate the quadrics, but repeat it four times. Here's what the code looks like, which then calculates the plane equations (the remaining parts are omitted for brevity):unsignedint i00 = indices[(i + 0) * 3 + 0];
unsignedint i01 = indices[(i + 0) * 3 + 1];
unsignedint i02 = indices[(i + 0) * 3 + 2];
unsignedint i10 = indices[(i + 1) * 3 + 0];
unsignedint i11 = indices[(i + 1) * 3 + 1];
unsignedint i12 = indices[(i + 1) * 3 + 2];
unsignedint i20 = indices[(i + 2) * 3 + 0];
unsignedint i21 = indices[(i + 2) * 3 + 1];
unsignedint i22 = indices[(i + 2) * 3 + 2];
unsignedint i30 = indices[(i + 3) * 3 + 0];
unsignedint i31 = indices[(i + 3) * 3 + 1];
unsignedint i32 = indices[(i + 3) * 3 + 2];
// load first vertex of each triangle and transpose into vectors with components (pw0 isn't used later)
__m128 px0 = _mm_loadu_ps(&vertex_positions[i00].x);
__m128 py0 = _mm_loadu_ps(&vertex_positions[i10].x);
__m128 pz0 = _mm_loadu_ps(&vertex_positions[i20].x);
__m128 pw0 = _mm_loadu_ps(&vertex_positions[i30].x);
_MM_TRANSPOSE4_PS(px0, py0, pz0, pw0);
// load second vertex of each triangle and transpose into vectors with components
__m128 px1 = _mm_loadu_ps(&vertex_positions[i01].x);
__m128 py1 = _mm_loadu_ps(&vertex_positions[i11].x);
__m128 pz1 = _mm_loadu_ps(&vertex_positions[i21].x);
__m128 pw1 = _mm_loadu_ps(&vertex_positions[i31].x);
_MM_TRANSPOSE4_PS(px1, py1, pz1, pw1);
// load third vertex of each triangle and transpose into vectors with components
__m128 px2 = _mm_loadu_ps(&vertex_positions[i02].x);
__m128 py2 = _mm_loadu_ps(&vertex_positions[i12].x);
__m128 pz2 = _mm_loadu_ps(&vertex_positions[i22].x);
__m128 pw2 = _mm_loadu_ps(&vertex_positions[i32].x);
_MM_TRANSPOSE4_PS(px2, py2, pz2, pw2);
// p1 - p0
__m128 px10 = _mm_sub_ps(px1, px0);
__m128 py10 = _mm_sub_ps(py1, py0);
__m128 pz10 = _mm_sub_ps(pz1, pz0);
// p2 - p0
__m128 px20 = _mm_sub_ps(px2, px0);
__m128 py20 = _mm_sub_ps(py2, py0);
__m128 pz20 = _mm_sub_ps(pz2, pz0);
// cross(p10, p20)
__m128 normalx = _mm_sub_ps(
_mm_mul_ps(py10, pz20),
_mm_mul_ps(pz10, py20));
__m128 normaly = _mm_sub_ps(
_mm_mul_ps(pz10, px20),
_mm_mul_ps(px10, pz20));
__m128 normalz = _mm_sub_ps(
_mm_mul_ps(px10, py20),
_mm_mul_ps(py10, px20));
// normalize; note that areasq/area now contain 4 values, not just one
__m128 areasq = _mm_add_ps(
_mm_mul_ps(normalx, normalx),
_mm_add_ps(
_mm_mul_ps(normaly, normaly),
_mm_mul_ps(normalz, normalz)));
__m128 area = _mm_sqrt_ps(areasq);
__m128 areanz = _mm_cmpneq_ps(area, _mm_setzero_ps());
normalx = _mm_and_ps(_mm_div_ps(normalx, area), areanz);
normaly = _mm_and_ps(_mm_div_ps(normaly, area), areanz);
normalz = _mm_and_ps(_mm_div_ps(normalz, area), areanz);
__m128 distance = _mm_add_ps(
_mm_mul_ps(normalx, px0),
_mm_add_ps(
_mm_mul_ps(normaly, py0),
_mm_mul_ps(normalz, pz0)));
__m128 negdistance = _mm_sub_ps(_mm_setzero_ps(), distance);
// this computes the plane equations (a, b, c, d) for each of the 4 triangles
__m128 plane0 = normalx;
__m128 plane1 = normaly;
__m128 plane2 = normalz;
__m128 plane3 = negdistance;
_MM_TRANSPOSE4_PS(plane0, plane1, plane2, plane3);The code has become a little longer: now we process four triangles in each iteration, and we no longer need SSE4.1 instructions for this. In theory, SIMD units should be used more efficiently. Let's see how it helped.
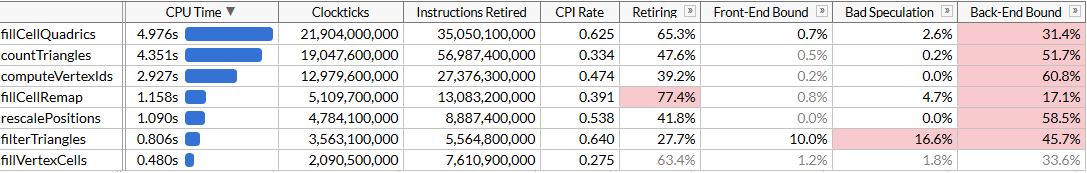
Okay, that's okay. The code has accelerated very slightly, although the function
fillCellQuadricsit now runs almost twice as fast as the original function without SIMD, but it is unclear whether this justifies a significant increase in complexity. Theoretically, you can use AVX2 and process 8 triangles per iteration, but here you will need to further spin the loop manually (ideally, all this code is generated using ISPC, but my naive attempts to get it to generate good code did not succeed: instead of the load / store sequences he persistently issued a gather / scatter, which significantly slows down the execution). Let's try something else.AVX2 = SSE2 + SSE2
The AVX2 is a slightly idiosyncratic set of instructions. It has 8-wide floating point registers, and 8 operations can be performed with one instruction; but in fact, such an instruction does not differ from two SSE2 instructions executed on two halves of the register (as far as I understand, the first processors with AVX2 support decoded each instruction in two or more microoperations, therefore the performance gain was limited by the phase of extracting instructions). For example, it
_mm_dp_psimplements a scalar product between two SSE2 registers, and _mm256_dp_psproduces two scalar products between two halves of two AVX2 registers, therefore it is limited to 4-wide for each half.Because of this, the AVX2 code often differs from the universal “8-wide SIMD”, but here it works in our favor: instead of trying to improve vectorization by transposing 4-wide vectors, we return to the first version of SIMD and double the loop using AVX2 instructions instead of SSE2 / SSE4. We still need to load and store 4-wide vectors, but in general we just change the code
__m128on __m256and _mm_on _mm256with a few settings:unsignedint i00 = indices[(i + 0) * 3 + 0];
unsignedint i01 = indices[(i + 0) * 3 + 1];
unsignedint i02 = indices[(i + 0) * 3 + 2];
unsignedint i10 = indices[(i + 1) * 3 + 0];
unsignedint i11 = indices[(i + 1) * 3 + 1];
unsignedint i12 = indices[(i + 1) * 3 + 2];
__m256 p0 = _mm256_loadu2_m128(
&vertex_positions[i10].x,
&vertex_positions[i00].x);
__m256 p1 = _mm256_loadu2_m128(
&vertex_positions[i11].x,
&vertex_positions[i01].x);
__m256 p2 = _mm256_loadu2_m128(
&vertex_positions[i12].x,
&vertex_positions[i02].x);
__m256 p10 = _mm256_sub_ps(p1, p0);
__m256 p20 = _mm256_sub_ps(p2, p0);
__m256 normal = _mm256_sub_ps(
_mm256_mul_ps(
_mm256_shuffle_ps(p10, p10, yzx),
_mm256_shuffle_ps(p20, p20, zxy)),
_mm256_mul_ps(
_mm256_shuffle_ps(p10, p10, zxy),
_mm256_shuffle_ps(p20, p20, yzx)));
__m256 areasq = _mm256_dp_ps(normal, normal, dp_xyz);
__m256 area = _mm256_sqrt_ps(areasq);
__m256 areanz = _mm256_cmp_ps(area, _mm256_setzero_ps(), _CMP_NEQ_OQ);
normal = _mm256_and_ps(_mm256_div_ps(normal, area), areanz);
__m256 distance = _mm256_dp_ps(normal, p0, dp_xyz);
__m256 negdistance = _mm256_sub_ps(_mm256_setzero_ps(), distance);
__m256 normalnegdist = _mm256_blend_ps(normal, negdistance, 0x88);
__m256 Qx = _mm256_mul_ps(normal, normal);
__m256 Qy = _mm256_mul_ps(
_mm256_shuffle_ps(normal, normal, _MM_SHUFFLE(3, 2, 2, 1)),
_mm256_shuffle_ps(normal, normal, _MM_SHUFFLE(3, 0, 1, 0)));
__m256 Qz = _mm256_mul_ps(negdistance, normalnegdist);Here you can take each 128-bit half of the received vectors
Qx/ Qy/ Qzand run the same code that we used to add the quadrics. Instead, we assume that if a triangle has three vertices in one cell (value single_cell == true), then it is very likely that another triangle has three vertices in another cell, and we perform the final aggregation of the quadrics using AVX2 as well:unsignedint c00 = vertex_cells[i00];
unsignedint c01 = vertex_cells[i01];
unsignedint c02 = vertex_cells[i02];
unsignedint c10 = vertex_cells[i10];
unsignedint c11 = vertex_cells[i11];
unsignedint c12 = vertex_cells[i12];
bool single_cell =
(c00 == c01) & (c00 == c02) &
(c10 == c11) & (c10 == c12);
if (single_cell)
{
area = _mm256_mul_ps(area, _mm256_set1_ps(3.f));
Qx = _mm256_mul_ps(Qx, area);
Qy = _mm256_mul_ps(Qy, area);
Qz = _mm256_mul_ps(Qz, area);
Quadric& q00 = cell_quadrics[c00];
Quadric& q10 = cell_quadrics[c10];
__m256 q0x = _mm256_loadu2_m128(&q10.a00, &q00.a00);
__m256 q0y = _mm256_loadu2_m128(&q10.a10, &q00.a10);
__m256 q0z = _mm256_loadu2_m128(&q10.b0, &q00.b0);
_mm256_storeu2_m128(&q10.a00, &q00.a00, _mm256_add_ps(q0x, Qx));
_mm256_storeu2_m128(&q10.a10, &q00.a10, _mm256_add_ps(q0y, Qy));
_mm256_storeu2_m128(&q10.b0, &q00.b0, _mm256_add_ps(q0z, Qz));
}
else
{
// omitted for brevity
}The resulting code is simpler, concise, and faster than our unsuccessful SSE2 approach:
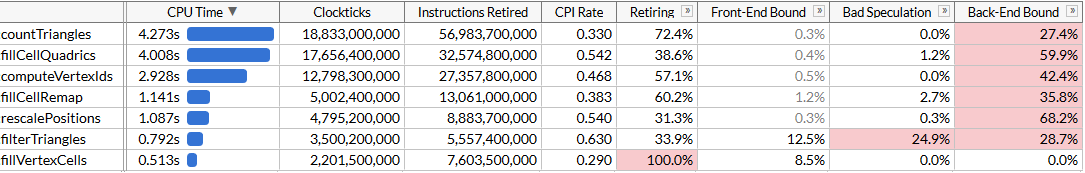
Of course, we did not achieve acceleration by 8 times, but only 2.45 times. The load and storage operations are still 4-wide, since we are forced to work with an inconvenient memory layout due to dynamic indexing, and the calculations are not optimal for SIMD. But now it’s
fillCellQuadricsno longer a bottleneck in the pipeline of our profile, and you can focus on other functions.Gather around, kids
We saved 4.8 seconds on the test run (48 ms on each run), and now our main intruder is
countTriangles. It would seem to be a simple function, but it is executed not once, but five times:static size_t countTriangles(constunsignedint* vertex_ids, constunsignedint* indices, size_t index_count){
size_t result = 0;
for (size_t i = 0; i < index_count; i += 3)
{
unsignedint id0 = vertex_ids[indices[i + 0]];
unsignedint id1 = vertex_ids[indices[i + 1]];
unsignedint id2 = vertex_ids[indices[i + 2]];
result += (id0 != id1) & (id0 != id2) & (id1 != id2);
}
return result;
}The function iterates over all the source triangles and calculates the number of non-degenerate triangles by comparing the identifiers of the vertices. It’s not immediately clear how to parallelize it using SIMD ... unless you use the gather instructions.
The AVX2 instruction set has added the gather / scatter instruction family to the x64 SIMD; each of them takes a vector register with 4 or 8 values - and simultaneously performs 4 or 8 load or save operations. If you use gather here, you can download three indexes, run gather for all at once (or in groups of 4 or 8) and compare the results. Gather on Intel processors has historically been pretty slow, but let's try it. For simplicity, we upload data for 8 triangles, transpose vectors in the same way as our previous attempt, and compare the corresponding elements of each vector:
for (size_t i = 0; i < (triangle_count & ~7); i += 8)
{
__m256 tri0 = _mm256_loadu2_m128(
(constfloat*)&indices[(i + 4) * 3 + 0],
(constfloat*)&indices[(i + 0) * 3 + 0]);
__m256 tri1 = _mm256_loadu2_m128(
(constfloat*)&indices[(i + 5) * 3 + 0],
(constfloat*)&indices[(i + 1) * 3 + 0]);
__m256 tri2 = _mm256_loadu2_m128(
(constfloat*)&indices[(i + 6) * 3 + 0],
(constfloat*)&indices[(i + 2) * 3 + 0]);
__m256 tri3 = _mm256_loadu2_m128(
(constfloat*)&indices[(i + 7) * 3 + 0],
(constfloat*)&indices[(i + 3) * 3 + 0]);
_MM_TRANSPOSE8_LANE4_PS(tri0, tri1, tri2, tri3);
__m256i i0 = _mm256_castps_si256(tri0);
__m256i i1 = _mm256_castps_si256(tri1);
__m256i i2 = _mm256_castps_si256(tri2);
__m256i id0 = _mm256_i32gather_epi32((int*)vertex_ids, i0, 4);
__m256i id1 = _mm256_i32gather_epi32((int*)vertex_ids, i1, 4);
__m256i id2 = _mm256_i32gather_epi32((int*)vertex_ids, i2, 4);
__m256i deg = _mm256_or_si256(
_mm256_cmpeq_epi32(id1, id2),
_mm256_or_si256(
_mm256_cmpeq_epi32(id0, id1),
_mm256_cmpeq_epi32(id0, id2)));
result += 8 - _mm_popcnt_u32(_mm256_movemask_epi8(deg)) / 4;
}A macro
_MM_TRANSPOSE8_LANE4_PSin AVX2 is the equivalent _MM_TRANSPOSE4_PSthat is missing from the standard header, but is easily output. It takes four AVX2 vectors and transposes two 4 × 4 matrices independently of each other:#define _MM_TRANSPOSE8_LANE4_PS(row0, row1, row2, row3) \
do { \
__m256 __t0, __t1, __t2, __t3; \
__t0 = _mm256_unpacklo_ps(row0, row1); \
__t1 = _mm256_unpackhi_ps(row0, row1); \
__t2 = _mm256_unpacklo_ps(row2, row3); \
__t3 = _mm256_unpackhi_ps(row2, row3); \
row0 = _mm256_shuffle_ps(__t0, __t2, _MM_SHUFFLE(1, 0, 1, 0)); \
row1 = _mm256_shuffle_ps(__t0, __t2, _MM_SHUFFLE(3, 2, 3, 2)); \
row2 = _mm256_shuffle_ps(__t1, __t3, _MM_SHUFFLE(1, 0, 1, 0)); \
row3 = _mm256_shuffle_ps(__t1, __t3, _MM_SHUFFLE(3, 2, 3, 2)); \
} while (0)Due to some features in the SSE2 / AVX2 instruction sets, we must use floating point register operations when transposing vectors. We are loading the data a little carelessly; but this basically doesn’t matter, because gather performance limits us:
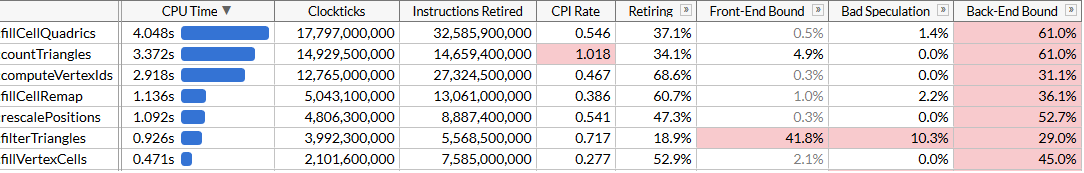
Now it
countTrianglesworks about 27% faster, and pay attention to the terrible CPI (loops per instruction): we send out about four times less instructions, but gather takes a lot of time. It's great that it speeds up the overall work, but, of course, the performance gain is somewhat depressing. We managed to overtake fillCellQuadricsin the profile, which brings us to the last function at the top of the list, which we have not looked at yet.Chapter 6, where everything starts to work as it should
The last function is this
computeVertexIds. In the algorithm, it is performed 6 times, therefore it also represents an excellent goal for optimization. For the first time, we see a function that seems to be created for clear optimization in SIMD:staticvoidcomputeVertexIds(unsignedint* vertex_ids, const Vector3* vertex_positions, size_t vertex_count, int grid_size){
assert(grid_size >= 1 && grid_size <= 1024);
float cell_scale = float(grid_size - 1);
for (size_t i = 0; i < vertex_count; ++i)
{
const Vector3& v = vertex_positions[i];
int xi = int(v.x * cell_scale + 0.5f);
int yi = int(v.y * cell_scale + 0.5f);
int zi = int(v.z * cell_scale + 0.5f);
vertex_ids[i] = (xi << 20) | (yi << 10) | zi;
}
}After the previous optimizations, we know what to do: unroll the cycle 4 or 8 times, since there is no point in trying to speed up only one iteration, transpose the vector components and start the calculations in parallel. We do this using AVX2, processing 8 vertices at a time:
__m256 scale = _mm256_set1_ps(cell_scale);
__m256 half = _mm256_set1_ps(0.5f);
for (size_t i = 0; i < (vertex_count & ~7); i += 8)
{
__m256 vx = _mm256_loadu2_m128(
&vertex_positions[i + 4].x,
&vertex_positions[i + 0].x);
__m256 vy = _mm256_loadu2_m128(
&vertex_positions[i + 5].x,
&vertex_positions[i + 1].x);
__m256 vz = _mm256_loadu2_m128(
&vertex_positions[i + 6].x,
&vertex_positions[i + 2].x);
__m256 vw = _mm256_loadu2_m128(
&vertex_positions[i + 7].x,
&vertex_positions[i + 3].x);
_MM_TRANSPOSE8_LANE4_PS(vx, vy, vz, vw);
__m256i xi = _mm256_cvttps_epi32(
_mm256_add_ps(_mm256_mul_ps(vx, scale), half));
__m256i yi = _mm256_cvttps_epi32(
_mm256_add_ps(_mm256_mul_ps(vy, scale), half));
__m256i zi = _mm256_cvttps_epi32(
_mm256_add_ps(_mm256_mul_ps(vz, scale), half));
__m256i id = _mm256_or_si256(
zi,
_mm256_or_si256(
_mm256_slli_epi32(xi, 20),
_mm256_slli_epi32(yi, 10)));
_mm256_storeu_si256((__m256i*)&vertex_ids[i], id);
}And look at the results:
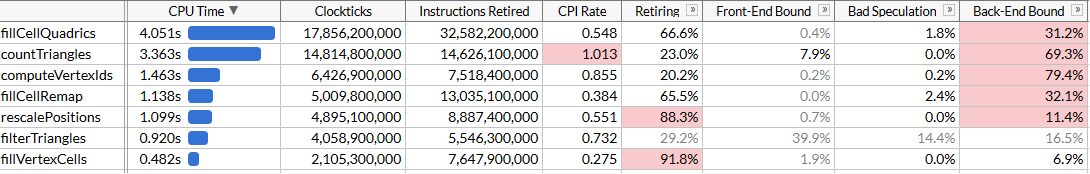
We accelerated
computeVertexIdstwice. Taking into account all the optimizations, the total execution time of the program was reduced to approximately 120 ms, which corresponds to the calculation of 50 million triangles per second. It may seem that we again did not achieve the expected increase in productivity: should
computeVertexIdsn't it accelerate more than twice after parallelization? To answer this question, let's try to see how much work this function performs. computeVertexIdsit is executed six times for one program start: five times during binary search and once at the end to calculate the final identifiers that are used for further processing. Each time this function processes 3 million vertices, reading 12 bytes for each vertex and writing 4 bytes.In total, over 100 runs of the innovator, this function processes 1.8 billion vertices, reading 21 GB and writing back 7 GB. Processing 28 GB in 1.46 seconds requires a bus bandwidth of 19 GB / s. We can check the memory bandwidth by running
memcmp(block1, block2, 512 MB). The result is 45 ms, that is, about 22 GB / s on one core (although the AIDA64 benchmark shows read speeds on my system up to 31 GB / s, but it uses several cores). In fact, we have come close to the maximum achievable memory limit, and a further increase in performance will require closer packing of these vertices so that they occupy less than 12 bytes.Conclusion
We took a fairly well-optimized algorithm that simplifies very large grids at a speed of 28 million triangles per second, and used the SSE and AVX instruction sets to speed it up almost twice, to 50 million triangles per second. During this journey, we had to learn different ways to use SIMD: registers for storing 3-wide vectors, SoA transposition, AVX2 instructions for storing two 3-wide vectors, gather instructions to speed up data loading compared to scalar instructions, and finally we directly applied AVX2 for streaming processing.
SIMD is often not the best starting point for optimization: the mesh optimizer went through many iterations of algorithmic optimization and microoptimization without using platform-specific instructions. But at some point, these possibilities are exhausted, and if performance is critical, then SIMD is a fantastic tool that can be used if necessary.
I’m not sure which of these optimizations will fall into the main branch
meshoptimizer: in the end, this is just an experiment to see how much the code accelerates without a radical change in the algorithms. I hope the article turned out to be informative and will give you ideas for optimizing the code. The final sources for this article are here ; this work is based on the version of meshoptimizer 99ab49, and the Thai Buddha model is published on Sketchfab .