Copying data from a website using R and the rvest library
To analyze data, you must first collect this data. There are many different methods for this purpose. In this article, we will talk about copying data directly from a website, or about scraping. There are several articles on Habr on how to make copying using Python. We will use the R language (ver. 3.4.2) and its rvest library. As an example, consider copying data from Google Scholar (GS).
GS is a search engine that does not search for information all over the Internet, but only in published articles or patents. This can be very helpful. For example, when searching for scientific articles for some keywords. To do this, enter these words in the search bar. Or, let's say you need to find articles published by a specific author. To do this, you can simply type his last name, but it is better to use the keyword 'author', and enter something like 'author: "D Smith"'.
Let's do this search and look at the result.
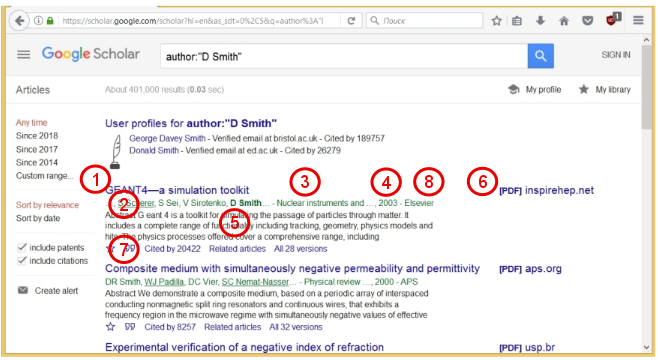
GS shows that about 400 thousand articles were found. For each article, its title (1), the names of the authors (2), the name of the journal (3), the year of issue (4), and a brief summary (5) are given. If a PDF file of the article is available, then the corresponding link (6) is given on the right. Also, an important parameter (7) is indicated for each article, namely, how many times this article was mentioned in other works ("Cited by"). This parameter shows how much this work is in demand. Using this parameter for several articles, you can evaluate the productivity and "demand" of the scientist. This is the so-called H-index of the scientist. According to Wikipedia , the Hirsch index is defined as follows:
"A scientist has index h if h from his Np articles is cited at least h times each, while the remaining (Np - h) articles are cited no more than h times each."
In other words, if the list of all articles of a certain author is ordered in decreasing order of number of citations then the serial number last article for which the condition , and there is a Hirsch index. Due to the fact that various scientific foundations and organizations now pay special attention to the Hirsch index and other scientometric indicators, people have learned to "wind up" these indicators. But that's another story.
In this article, we will search for articles by a specific author. For example, take the Russian scientist Alexei Yakovlevich Chervonenkis . A. Ya. Chervonenkis, a well-known Russian scientist in the field of computer science, made a significant contribution to the theory of data analysis and machine learning. He died tragically in 2014.
We will collect the following 7 parameters of the articles: Title of the article (1), List of authors (2), Title of the journal (3), Year of issue (4), Publisher (8), Number of citations (7), Link to citing articles (7) . The last parameter is needed, for example, if we want to see the relationship of the authors, to identify the network of people conducting research in a certain direction.
Training
First you need to install the rvest package. To do this, use the following R command:
install.packages(rvest)Now we need a tool that shows which component of the web page code corresponds to one or another parameter. You can use the tools built into the browser. For example, in Mozilla, you can select from the Menu: "Tools -> Web Development -> Inspector". The HTML code of the web page will be displayed. Then, hovering over some element of the page, you can see which CSS code corresponds to it. But we will do it easier. We will use the SelectorGadget tool . To do this, go to the specified site and add a link to the code (located at the end of the program description) in the Bookmarks of your browser.

Now, being on any page, you can click on this tab, and a convenient tool will appear that allows you to determine the code components of this page (for details, see below).
Before copying the data, it is also useful to study the properties of the web page, namely, how the page changes from the type of request, to which addresses the various links correspond, etc. So, we will search for articles by A. Ya. Chervonenkis by the following query: author: "A Chervonenkis". This corresponds to the following address:
https://scholar.google.com/scholar?hl=en&as_sdt=1%2C5&as_vis=1&q=author%3A%22A+Chervonenkis%22&btnG=With this query syntax, patents are not taken into account, as well as articles that link to Chervonenkis, but in which he is not the author.
Copy data
Now let's get the data copying program. First, we connect the rvest library:
library(rvest)Next, set the required address and read the HTML-code of the web page:
url <- 'https://scholar.google.com/scholar?hl=en&as_sdt=1%2C5&as_vis=1&q=author%3A%22A+Chervonenkis%22&btnG='
wpage <- read_html(url)Now copy the names of the articles. To do this, run SelectorGadget, and click on the title of an article. This name is highlighted in green, but other components of the page are also highlighted (in yellow). The SelectorGadget line shows that 152 components were initially selected.
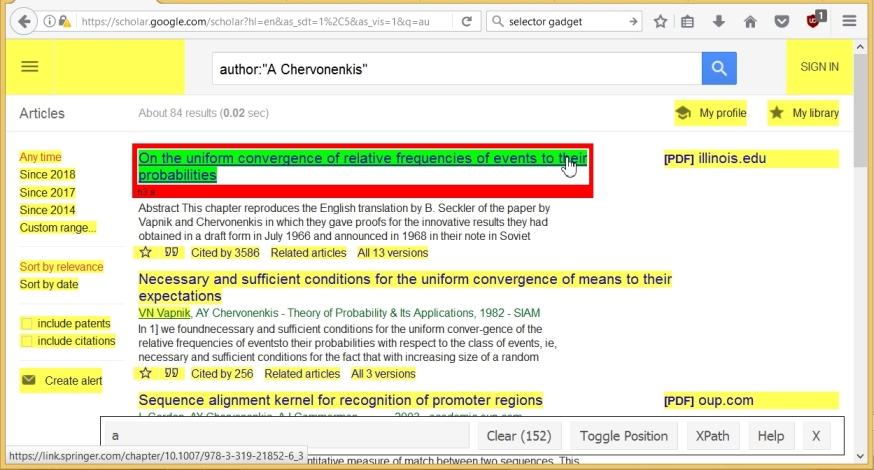
Now we just click on the components we don’t need. As a result, there are only 10 of them (according to the number of articles on the page), and the name of the component corresponding to the name of the article, namely ".gs_rt a", is given in the SelectorGadget'a line.
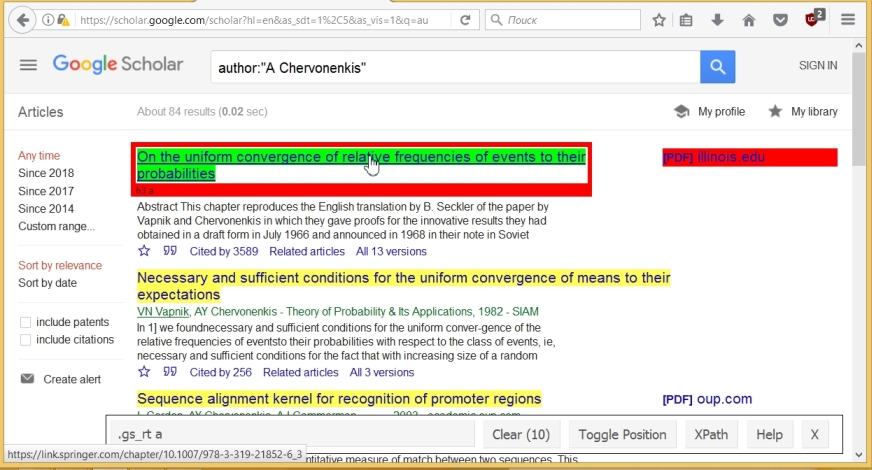
Using this name we can copy all the titles and convert them to text format using the following commands (the last command gives the structure of the titles variable ):
titles <- html_nodes(wpage, '.gs_rt a')
titles <- html_text(titles)
str(titles)
## chr [1:10] "On the uniform convergence of relative frequencies of ..."Hereinafter, after the characters '##', the program output (in truncated form) is shown.
Similarly, we determine that the names of the authors, the name of the journal, the year, and the publisher correspond to the component .gs_a. At the same time, the text of this component has the following format "<authors> - <journal, year> - <publisher>". We extract the text from ".gs_a", select the parameters we need ( authors , journals , year , publ ) in accordance with the format, remove unnecessary characters and spaces
# Scrap combined data, convert to text format:
comb <- html_nodes(wpage,'.gs_a')
comb <- html_text(comb)
str(comb)
## chr [1:10] "VN Vapnik, AY Chervonenkis - Measures of complexity, ..."
lst <- strsplit(comb, '-')
# Find authors, journal, year, publisher, extracting components of list
authors <- sapply(lst, '[[', 1) # Take 1st component of list
publ <- sapply(lst, '[[', 3)
lst1 <- strsplit( sapply(lst, '[[', 2), ',')
journals <- sapply(lst1, '[[', 1)
year <- sapply(lst1, '[[', 2)
# Replace 3 dots with ~, trim spaces, convert 'year' to numeric :
authors <- trimws(gsub(authors, pattern= '…', replacement= '~'))
journals <- trimws(gsub(journals, pattern= '…', replacement= '~'))
year <- as.numeric(gsub(year, pattern= '…', replacement= '~'))
publ <- trimws(gsub(publ, pattern= '…', replacement= '~'))
str(authors)
## chr [1:10] "VN Vapnik, AY Chervonenkis " "VN Vapnik, AY Chervonenkis " ...Note that sometimes the name of the journal contains a hyphen "-". In this case, the procedure for extracting parameters from the source string will change somewhat.
Next, using SelectorGadget, determine the name of the component for the number of citations per article, remove unnecessary words, and convert the data to a number format
cit0 <- html_nodes(wpage,'#gs_res_ccl_mid a:nth-child(3)')
cit <- html_text(cit0)
lst <- strsplit(cit, ' ')
cit <- as.numeric(sapply(lst, '[[', 3))
str(cit)
## num [1:10] 3586 256 136 102 30 ...And finally, we extract the appropriate link:
cit_link <- html_attr(cit0, 'href')
str(cit_link)
## chr [1:10] "/scholar?cites=3657561935311739131&as_sdt=2005&..."Now we have 7 vectors ( titles , authors , journals , year , publ , cit , cit_link ) for our 7 parameters. We can combine them into a single structure (dataframe)
df1 <- data.frame(titles= titles, authors= authors,
journals= journals, year= year, publ = publ,
cit= cit, cit_link= cit_link, stringsAsFactors = FALSE)You can programmatically go to the next page by adding to the address 'start = n &', where n / 10 + 1 corresponds to the page number. Thus, it is possible to collect information on all articles of the
author. Further, using links to cited articles ( cit_link ), you can find data on other authors.
In conclusion, a few comments. The following is written in the Google Terms of Service :
"Don’t misuse our Services. For example, don’t interfere with our Services or try to access them using a method other than the interface and the instructions that we provide."
Информация в Интернете свидетельствует о том, что Гугл отслеживает доступ к его веб-страницам, в частности в GS. Если Гугл подозревает, что информация извлекается с помощью бота, он может ограничить или закрыть доступ к информации с определенного IP-адреса. Например, если запросы идут слишком часто, или через равные промежутки времени, то такое поведение рассматривается, как подозрительное.
Рассмотренный метод можно легко адаптировать к другим веб-сайтам. Комбинация R, rvest SelectorGadget делает копирование данных достаточно простым.
При подготовке данной статьи использовалась информация отсюда.
